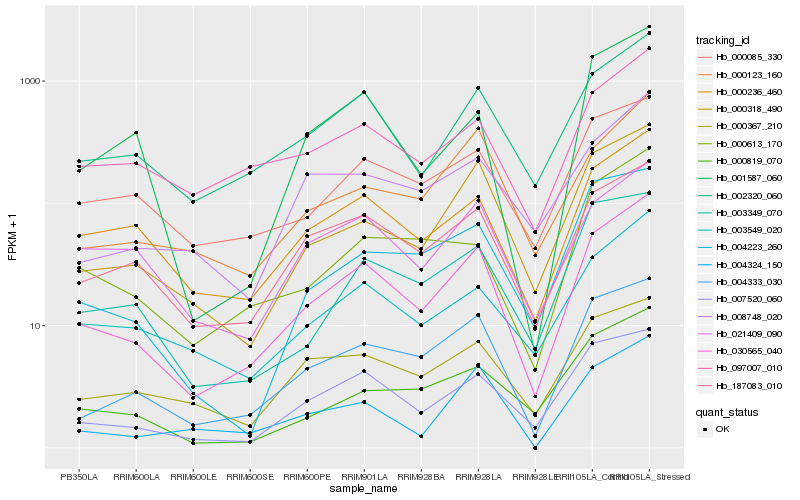
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_030565_040 |
0.0 |
- |
- |
glycine-rich RNA-binding protein, putative [Ricinus communis] |
| 2 |
Hb_004333_030 |
0.1044798989 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 26A [Jatropha curcas] |
| 3 |
Hb_007520_060 |
0.1081427776 |
- |
- |
PREDICTED: putative exosome complex component rrp40 [Jatropha curcas] |
| 4 |
Hb_002320_060 |
0.1083510006 |
- |
- |
hypothetical protein CARUB_v10018269mg, partial [Capsella rubella] |
| 5 |
Hb_000318_490 |
0.1130400979 |
- |
- |
PREDICTED: mitochondrial import receptor subunit TOM6 homolog [Jatropha curcas] |
| 6 |
Hb_187083_010 |
0.1174083067 |
- |
- |
PREDICTED: uncharacterized protein LOC105643971 [Jatropha curcas] |
| 7 |
Hb_000236_460 |
0.1208139858 |
- |
- |
PREDICTED: uncharacterized protein LOC105633119 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000123_160 |
0.1285436749 |
- |
- |
PREDICTED: 40S ribosomal protein S15a-1 [Jatropha curcas] |
| 9 |
Hb_003549_020 |
0.129504672 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_097007_010 |
0.1376944565 |
- |
- |
hypothetical protein CARUB_v10018269mg, partial [Capsella rubella] |
| 11 |
Hb_000085_330 |
0.1388020753 |
- |
- |
PREDICTED: 40S ribosomal protein S8 [Jatropha curcas] |
| 12 |
Hb_000367_210 |
0.1403123252 |
- |
- |
PREDICTED: dolichol-phosphate mannosyltransferase subunit 3 [Jatropha curcas] |
| 13 |
Hb_004324_150 |
0.1404220822 |
- |
- |
- |
| 14 |
Hb_003349_070 |
0.1410007359 |
- |
- |
PREDICTED: signal recognition particle 14 kDa protein [Gossypium raimondii] |
| 15 |
Hb_000819_070 |
0.1420396192 |
- |
- |
PREDICTED: dolichol-phosphate mannosyltransferase subunit 1 [Jatropha curcas] |
| 16 |
Hb_000613_170 |
0.1423645951 |
- |
- |
PREDICTED: RPM1-interacting protein 4 [Jatropha curcas] |
| 17 |
Hb_021409_090 |
0.1425207802 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0003s14450g [Populus trichocarpa] |
| 18 |
Hb_008748_020 |
0.1437946751 |
- |
- |
hypothetical protein B456_005G210700 [Gossypium raimondii] |
| 19 |
Hb_001587_060 |
0.1441637325 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_004223_260 |
0.1445019541 |
- |
- |
hypothetical protein F383_09277 [Gossypium arboreum] |