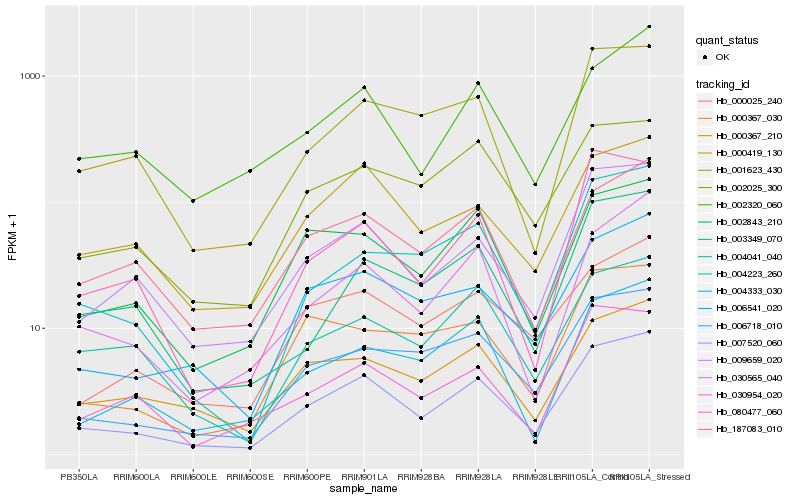
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007520_060 |
0.0 |
- |
- |
PREDICTED: putative exosome complex component rrp40 [Jatropha curcas] |
| 2 |
Hb_009659_020 |
0.0956660471 |
- |
- |
PREDICTED: uncharacterized protein LOC103870090 [Brassica rapa] |
| 3 |
Hb_006541_020 |
0.1066767882 |
- |
- |
unknown [Lotus japonicus] |
| 4 |
Hb_030565_040 |
0.1081427776 |
- |
- |
glycine-rich RNA-binding protein, putative [Ricinus communis] |
| 5 |
Hb_006718_010 |
0.1140374191 |
- |
- |
PREDICTED: histone H2AX-like [Elaeis guineensis] |
| 6 |
Hb_000367_210 |
0.1183037543 |
- |
- |
PREDICTED: dolichol-phosphate mannosyltransferase subunit 3 [Jatropha curcas] |
| 7 |
Hb_000419_130 |
0.1198014075 |
- |
- |
PREDICTED: uncharacterized protein LOC105650901 [Jatropha curcas] |
| 8 |
Hb_001623_430 |
0.1205770339 |
- |
- |
PREDICTED: small acidic protein 1 [Jatropha curcas] |
| 9 |
Hb_004223_260 |
0.1211055799 |
- |
- |
hypothetical protein F383_09277 [Gossypium arboreum] |
| 10 |
Hb_000025_240 |
0.1212167207 |
- |
- |
PREDICTED: uncharacterized protein LOC105628964 [Jatropha curcas] |
| 11 |
Hb_002843_210 |
0.121402598 |
- |
- |
PREDICTED: uncharacterized protein LOC105647178 [Jatropha curcas] |
| 12 |
Hb_187083_010 |
0.1216132673 |
- |
- |
PREDICTED: uncharacterized protein LOC105643971 [Jatropha curcas] |
| 13 |
Hb_003349_070 |
0.1268034152 |
- |
- |
PREDICTED: signal recognition particle 14 kDa protein [Gossypium raimondii] |
| 14 |
Hb_004333_030 |
0.1272516346 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 26A [Jatropha curcas] |
| 15 |
Hb_002320_060 |
0.1282034624 |
- |
- |
hypothetical protein CARUB_v10018269mg, partial [Capsella rubella] |
| 16 |
Hb_002025_300 |
0.1282422533 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 7 [Jatropha curcas] |
| 17 |
Hb_004041_040 |
0.1290802851 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM14-1 [Camelina sativa] |
| 18 |
Hb_080477_060 |
0.1308218676 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_030954_020 |
0.1330987148 |
- |
- |
hypothetical protein RCOM_1330200 [Ricinus communis] |
| 20 |
Hb_000367_030 |
0.1369871298 |
- |
- |
hypothetical protein POPTR_0015s06690g [Populus trichocarpa] |