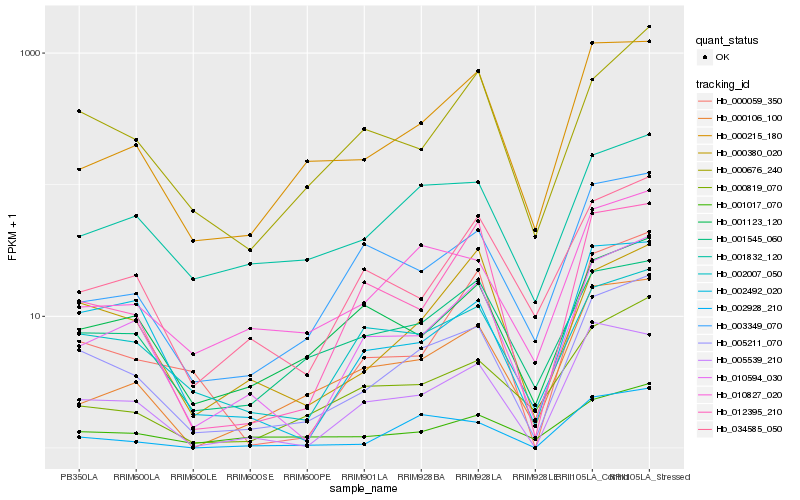
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005211_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642009 [Jatropha curcas] |
| 2 |
Hb_010594_030 |
0.1294567696 |
- |
- |
- |
| 3 |
Hb_002492_020 |
0.1448478077 |
- |
- |
PREDICTED: uncharacterized protein LOC104789613 [Camelina sativa] |
| 4 |
Hb_010827_020 |
0.1485782651 |
- |
- |
60S ribosomal protein L17B [Hevea brasiliensis] |
| 5 |
Hb_000215_180 |
0.1494706344 |
- |
- |
hypothetical protein 17 [Hevea brasiliensis] |
| 6 |
Hb_005539_210 |
0.1507579658 |
- |
- |
- |
| 7 |
Hb_000676_240 |
0.1510271111 |
- |
- |
thioredoxin H-type 2 [Hevea brasiliensis] |
| 8 |
Hb_002007_050 |
0.1555158443 |
- |
- |
PREDICTED: putative protease Do-like 14 [Jatropha curcas] |
| 9 |
Hb_002928_210 |
0.1569305102 |
- |
- |
- |
| 10 |
Hb_001545_060 |
0.161155824 |
- |
- |
PREDICTED: uncharacterized protein LOC105643880 [Jatropha curcas] |
| 11 |
Hb_001017_070 |
0.1619964889 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000380_020 |
0.1626600301 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger protein 36, C3H1 type-like 3 [Jatropha curcas] |
| 13 |
Hb_000106_100 |
0.1632907019 |
- |
- |
- |
| 14 |
Hb_012395_210 |
0.1637237406 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000059_350 |
0.1644791362 |
- |
- |
glutamate dehydrogenase, putative [Ricinus communis] |
| 16 |
Hb_000819_070 |
0.1646931186 |
- |
- |
PREDICTED: dolichol-phosphate mannosyltransferase subunit 1 [Jatropha curcas] |
| 17 |
Hb_001832_120 |
0.1654422153 |
- |
- |
hypothetical protein PHAVU_003G136800g [Phaseolus vulgaris] |
| 18 |
Hb_001123_120 |
0.1655579127 |
- |
- |
Uncharacterized protein TCM_042776 [Theobroma cacao] |
| 19 |
Hb_003349_070 |
0.1661835369 |
- |
- |
PREDICTED: signal recognition particle 14 kDa protein [Gossypium raimondii] |
| 20 |
Hb_034585_050 |
0.1680276162 |
- |
- |
- |