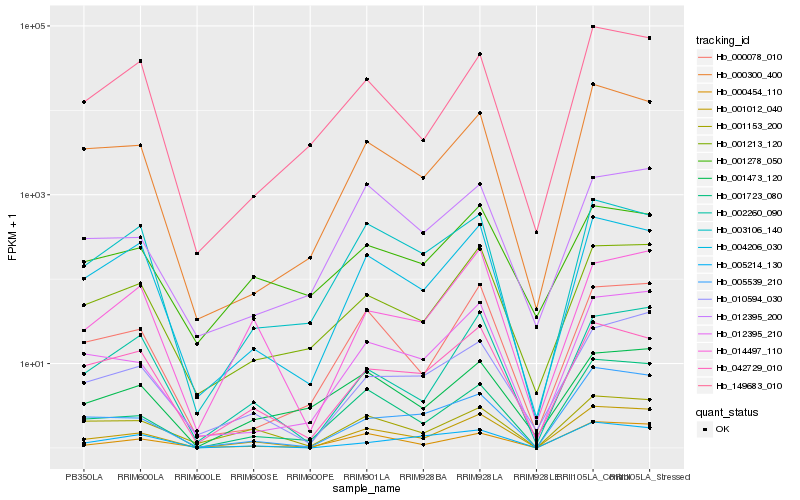
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001012_040 |
0.0 |
- |
- |
PREDICTED: MADS-box transcription factor 23-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_001723_080 |
0.1183187746 |
- |
- |
hypothetical protein L484_026139 [Morus notabilis] |
| 3 |
Hb_000454_110 |
0.1306350196 |
- |
- |
PREDICTED: polyprenol reductase 2-like isoform X3 [Populus euphratica] |
| 4 |
Hb_012395_210 |
0.1357939994 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_004206_030 |
0.1366521005 |
- |
- |
PREDICTED: protein indeterminate-domain 2-like [Jatropha curcas] |
| 6 |
Hb_005539_210 |
0.1371047574 |
- |
- |
- |
| 7 |
Hb_001213_120 |
0.1379662032 |
- |
- |
PREDICTED: immediate early response 3-interacting protein 1-like [Brassica rapa] |
| 8 |
Hb_000078_010 |
0.1453153833 |
- |
- |
hypothetical protein POPTR_0006s06510g, partial [Populus trichocarpa] |
| 9 |
Hb_000300_400 |
0.1460988551 |
- |
- |
hypothetical protein PHAVU_001G114100g [Phaseolus vulgaris] |
| 10 |
Hb_014497_110 |
0.1468746195 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 25-like [Jatropha curcas] |
| 11 |
Hb_010594_030 |
0.1507679396 |
- |
- |
- |
| 12 |
Hb_002260_090 |
0.1526091513 |
- |
- |
PREDICTED: uncharacterized protein LOC105636025 isoform X1 [Jatropha curcas] |
| 13 |
Hb_042729_010 |
0.1526912402 |
- |
- |
- |
| 14 |
Hb_001153_200 |
0.1533773687 |
- |
- |
PREDICTED: calcium-dependent protein kinase 26 isoform X1 [Jatropha curcas] |
| 15 |
Hb_003106_140 |
0.1612712229 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001278_050 |
0.1620454827 |
- |
- |
PREDICTED: uncharacterized protein LOC105636924 [Jatropha curcas] |
| 17 |
Hb_001473_120 |
0.1663568548 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 18 |
Hb_005214_130 |
0.1669559885 |
- |
- |
- |
| 19 |
Hb_012395_200 |
0.1710721733 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_149683_010 |
0.171918781 |
rubber biosynthesis |
Gene Name: REF2 |
RecName: Full=Rubber elongation factor protein; Short=REF; AltName: Allergen=Hev b 1 [Hevea brasiliensis] |