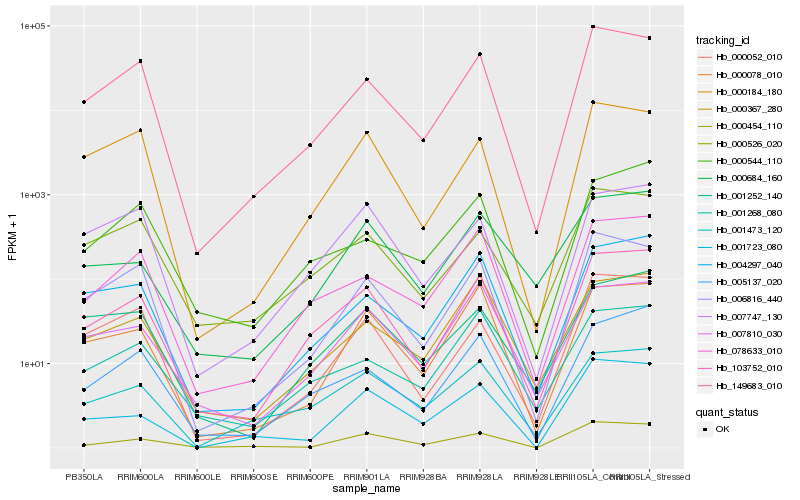
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_103752_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At5g39865-like [Jatropha curcas] |
| 2 |
Hb_005137_020 |
0.1062824565 |
- |
- |
PREDICTED: uncharacterized protein LOC101759641 [Setaria italica] |
| 3 |
Hb_149683_010 |
0.1124153068 |
rubber biosynthesis |
Gene Name: REF2 |
RecName: Full=Rubber elongation factor protein; Short=REF; AltName: Allergen=Hev b 1 [Hevea brasiliensis] |
| 4 |
Hb_078633_010 |
0.1161166403 |
- |
- |
PREDICTED: RNA-binding protein 24-B isoform X1 [Jatropha curcas] |
| 5 |
Hb_004297_040 |
0.1200239122 |
- |
- |
acyl CoA reductase [Hevea brasiliensis] |
| 6 |
Hb_000684_160 |
0.1271682914 |
- |
- |
PREDICTED: lipid phosphate phosphatase gamma, chloroplastic [Jatropha curcas] |
| 7 |
Hb_006816_440 |
0.1300724425 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 8 |
Hb_000052_010 |
0.1307829508 |
- |
- |
hypothetical protein M569_08416, partial [Genlisea aurea] |
| 9 |
Hb_001252_140 |
0.1325569393 |
- |
- |
catalytic, putative [Ricinus communis] |
| 10 |
Hb_000454_110 |
0.1328248101 |
- |
- |
PREDICTED: polyprenol reductase 2-like isoform X3 [Populus euphratica] |
| 11 |
Hb_001473_120 |
0.1360535705 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 12 |
Hb_000526_020 |
0.1361313899 |
- |
- |
thioredoxin-like family protein [Hevea brasiliensis] |
| 13 |
Hb_000078_010 |
0.1366754884 |
- |
- |
hypothetical protein POPTR_0006s06510g, partial [Populus trichocarpa] |
| 14 |
Hb_007810_030 |
0.1385427636 |
- |
- |
PREDICTED: uncharacterized protein LOC105647693 [Jatropha curcas] |
| 15 |
Hb_000184_180 |
0.1386665642 |
- |
- |
putative C3HC4-type RING zinc finger protein [Hevea brasiliensis] |
| 16 |
Hb_001268_080 |
0.1401667304 |
- |
- |
catalytic, putative [Ricinus communis] |
| 17 |
Hb_000544_110 |
0.1407951785 |
- |
- |
PREDICTED: probable calcium-binding protein CML27 [Jatropha curcas] |
| 18 |
Hb_000367_280 |
0.1424855495 |
- |
- |
PREDICTED: flavin-containing monooxygenase FMO GS-OX-like 9 [Jatropha curcas] |
| 19 |
Hb_007747_130 |
0.1444949824 |
- |
- |
PREDICTED: uncharacterized protein LOC105635879 [Jatropha curcas] |
| 20 |
Hb_001723_080 |
0.1445438132 |
- |
- |
hypothetical protein L484_026139 [Morus notabilis] |