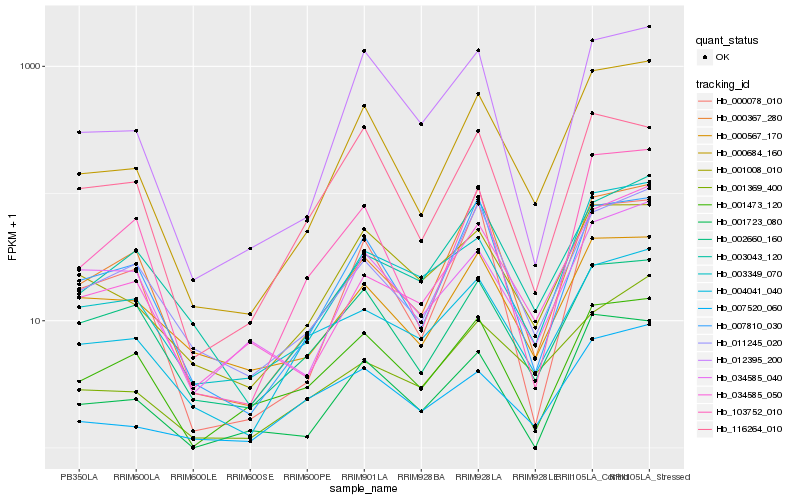
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000684_160 |
0.0 |
- |
- |
PREDICTED: lipid phosphate phosphatase gamma, chloroplastic [Jatropha curcas] |
| 2 |
Hb_012395_200 |
0.1219586002 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_003349_070 |
0.1252745558 |
- |
- |
PREDICTED: signal recognition particle 14 kDa protein [Gossypium raimondii] |
| 4 |
Hb_007810_030 |
0.1270442324 |
- |
- |
PREDICTED: uncharacterized protein LOC105647693 [Jatropha curcas] |
| 5 |
Hb_103752_010 |
0.1271682914 |
- |
- |
PREDICTED: uncharacterized protein At5g39865-like [Jatropha curcas] |
| 6 |
Hb_001008_010 |
0.1296243563 |
- |
- |
PREDICTED: 30S ribosomal protein S17, chloroplastic-like [Populus euphratica] |
| 7 |
Hb_011245_020 |
0.1364606015 |
- |
- |
Ferrochelatase [Medicago truncatula] |
| 8 |
Hb_034585_050 |
0.1382697 |
- |
- |
- |
| 9 |
Hb_000078_010 |
0.1407107272 |
- |
- |
hypothetical protein POPTR_0006s06510g, partial [Populus trichocarpa] |
| 10 |
Hb_001723_080 |
0.1426734792 |
- |
- |
hypothetical protein L484_026139 [Morus notabilis] |
| 11 |
Hb_004041_040 |
0.1430403949 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM14-1 [Camelina sativa] |
| 12 |
Hb_007520_060 |
0.1475906058 |
- |
- |
PREDICTED: putative exosome complex component rrp40 [Jatropha curcas] |
| 13 |
Hb_000367_280 |
0.1485195323 |
- |
- |
PREDICTED: flavin-containing monooxygenase FMO GS-OX-like 9 [Jatropha curcas] |
| 14 |
Hb_002660_160 |
0.1489635718 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 15 |
Hb_001473_120 |
0.1505759841 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 16 |
Hb_003043_120 |
0.1530668466 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_034585_040 |
0.1531746893 |
- |
- |
PREDICTED: nuclear pore complex protein NUP58 [Jatropha curcas] |
| 18 |
Hb_001369_400 |
0.1534316376 |
- |
- |
PREDICTED: uncharacterized protein LOC105648609 [Jatropha curcas] |
| 19 |
Hb_116264_010 |
0.1535593563 |
- |
- |
hypothetical protein JCGZ_05585 [Jatropha curcas] |
| 20 |
Hb_000567_170 |
0.1554741285 |
- |
- |
PREDICTED: putative lipase YDR444W isoform X1 [Jatropha curcas] |