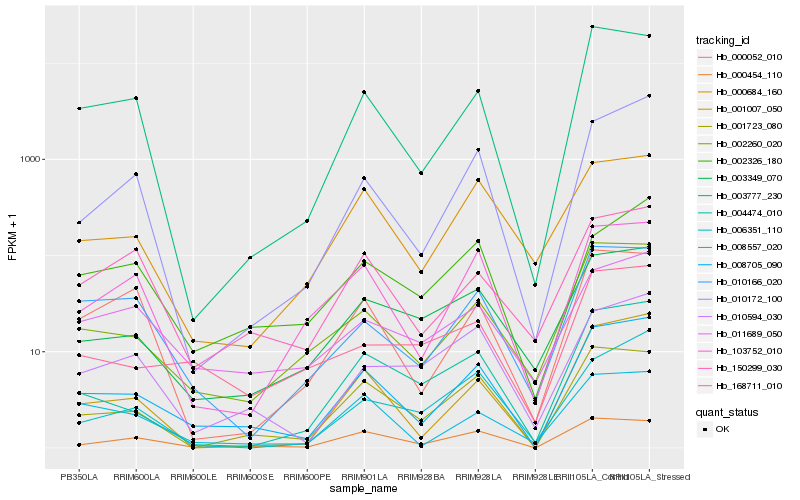
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008705_090 |
0.0 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-2-like [Jatropha curcas] |
| 2 |
Hb_001007_050 |
0.1285237495 |
- |
- |
hypothetical protein MIMGU_mgv1a0196712mg, partial [Erythranthe guttata] |
| 3 |
Hb_002260_020 |
0.1309392147 |
- |
- |
- |
| 4 |
Hb_001723_080 |
0.1323850432 |
- |
- |
hypothetical protein L484_026139 [Morus notabilis] |
| 5 |
Hb_003777_230 |
0.1394695636 |
- |
- |
PREDICTED: abscisic stress-ripening protein 1-like [Citrus sinensis] |
| 6 |
Hb_010172_100 |
0.1427874442 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26 [Populus euphratica] |
| 7 |
Hb_000454_110 |
0.1462093291 |
- |
- |
PREDICTED: polyprenol reductase 2-like isoform X3 [Populus euphratica] |
| 8 |
Hb_004474_010 |
0.1466989824 |
- |
- |
unnamed protein product [Coffea canephora] |
| 9 |
Hb_003349_070 |
0.1556583408 |
- |
- |
PREDICTED: signal recognition particle 14 kDa protein [Gossypium raimondii] |
| 10 |
Hb_103752_010 |
0.1589935905 |
- |
- |
PREDICTED: uncharacterized protein At5g39865-like [Jatropha curcas] |
| 11 |
Hb_010166_020 |
0.1597593376 |
transcription factor |
TF Family: NF-YC |
ccaat-binding transcription factor, putative [Ricinus communis] |
| 12 |
Hb_150299_030 |
0.1630417854 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_011689_050 |
0.1656133856 |
- |
- |
hypothetical protein POPTR_0008s20290g [Populus trichocarpa] |
| 14 |
Hb_008557_020 |
0.1667221085 |
- |
- |
PREDICTED: uncharacterized protein LOC105638665 [Jatropha curcas] |
| 15 |
Hb_000684_160 |
0.1669970022 |
- |
- |
PREDICTED: lipid phosphate phosphatase gamma, chloroplastic [Jatropha curcas] |
| 16 |
Hb_168711_010 |
0.1678628893 |
- |
- |
PREDICTED: (DL)-glycerol-3-phosphatase 2 [Sesamum indicum] |
| 17 |
Hb_010594_030 |
0.1679754941 |
- |
- |
- |
| 18 |
Hb_002326_180 |
0.1741224781 |
- |
- |
Protein Z, putative [Ricinus communis] |
| 19 |
Hb_006351_110 |
0.1744259295 |
- |
- |
- |
| 20 |
Hb_000052_010 |
0.1750260653 |
- |
- |
hypothetical protein M569_08416, partial [Genlisea aurea] |