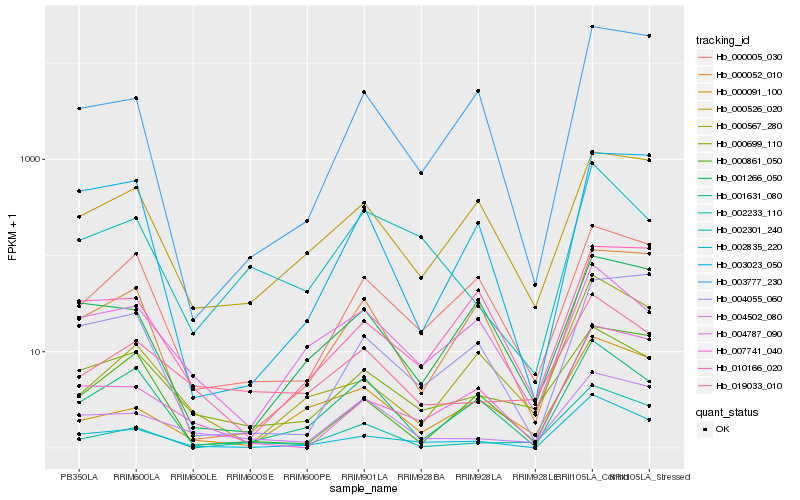
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002835_220 |
0.0 |
- |
- |
PREDICTED: MATE efflux family protein 7 [Vitis vinifera] |
| 2 |
Hb_007741_040 |
0.1697657215 |
transcription factor |
TF Family: PLATZ |
PREDICTED: uncharacterized protein LOC105643153 [Jatropha curcas] |
| 3 |
Hb_000567_280 |
0.185487657 |
- |
- |
PREDICTED: uncharacterized protein At4g06598-like [Populus euphratica] |
| 4 |
Hb_003777_230 |
0.1929289339 |
- |
- |
PREDICTED: abscisic stress-ripening protein 1-like [Citrus sinensis] |
| 5 |
Hb_000005_030 |
0.1936343732 |
- |
- |
hypothetical protein POPTR_0009s00870g [Populus trichocarpa] |
| 6 |
Hb_001631_080 |
0.1998802114 |
- |
- |
PREDICTED: importin-5-like [Populus euphratica] |
| 7 |
Hb_000526_020 |
0.2036442884 |
- |
- |
thioredoxin-like family protein [Hevea brasiliensis] |
| 8 |
Hb_002233_110 |
0.2042548312 |
- |
- |
PREDICTED: nuclear transcription factor Y subunit B-3-like [Jatropha curcas] |
| 9 |
Hb_000861_050 |
0.2061822051 |
- |
- |
PREDICTED: transcription elongation factor SPT6-like isoform X1 [Populus euphratica] |
| 10 |
Hb_002301_240 |
0.2069021155 |
- |
- |
auxin-repressed protein-like protein ARP1 [Manihot esculenta] |
| 11 |
Hb_019033_010 |
0.2070746608 |
- |
- |
PREDICTED: uncharacterized protein LOC105643397 [Jatropha curcas] |
| 12 |
Hb_001266_050 |
0.2073793493 |
- |
- |
PREDICTED: lipid phosphate phosphatase epsilon 2, chloroplastic-like isoform X2 [Elaeis guineensis] |
| 13 |
Hb_004502_080 |
0.2080628241 |
- |
- |
hypothetical protein JCGZ_23578 [Jatropha curcas] |
| 14 |
Hb_000052_010 |
0.2090871499 |
- |
- |
hypothetical protein M569_08416, partial [Genlisea aurea] |
| 15 |
Hb_000091_100 |
0.2096487258 |
- |
- |
PREDICTED: arginine biosynthesis bifunctional protein ArgJ, chloroplastic isoform X1 [Jatropha curcas] |
| 16 |
Hb_004787_090 |
0.2113445927 |
- |
- |
PREDICTED: HVA22-like protein a [Jatropha curcas] |
| 17 |
Hb_004055_060 |
0.2137461505 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 5 [Jatropha curcas] |
| 18 |
Hb_000699_110 |
0.2141115243 |
- |
- |
hypothetical protein POPTR_0013s04180g [Populus trichocarpa] |
| 19 |
Hb_003023_050 |
0.2148081812 |
- |
- |
hypothetical protein POPTR_0013s10320g [Populus trichocarpa] |
| 20 |
Hb_010166_020 |
0.2151856554 |
transcription factor |
TF Family: NF-YC |
ccaat-binding transcription factor, putative [Ricinus communis] |