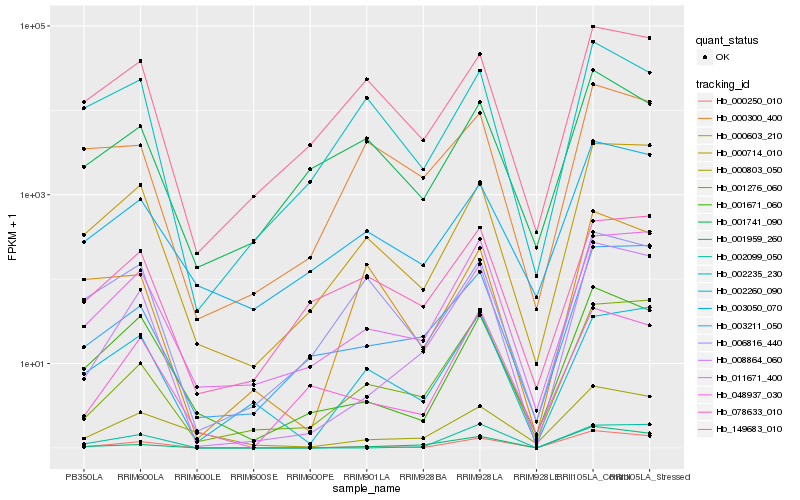
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000250_010 |
0.0 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 2 |
Hb_008864_060 |
0.1054710537 |
- |
- |
PREDICTED: uncharacterized protein LOC105634361 [Jatropha curcas] |
| 3 |
Hb_000714_010 |
0.1250982924 |
- |
- |
PREDICTED: salt stress-induced hydrophobic peptide ESI3 [Vitis vinifera] |
| 4 |
Hb_001671_060 |
0.1377505038 |
- |
- |
fad NAD binding oxidoreductases, putative [Ricinus communis] |
| 5 |
Hb_011671_400 |
0.1434009084 |
- |
- |
PREDICTED: cyclin-U4-1 [Jatropha curcas] |
| 6 |
Hb_001959_260 |
0.1490000227 |
- |
- |
- |
| 7 |
Hb_003050_070 |
0.1577426019 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_149683_010 |
0.1618521321 |
rubber biosynthesis |
Gene Name: REF2 |
RecName: Full=Rubber elongation factor protein; Short=REF; AltName: Allergen=Hev b 1 [Hevea brasiliensis] |
| 9 |
Hb_001276_060 |
0.1676999799 |
- |
- |
hypothetical protein POPTR_0015s00420g [Populus trichocarpa] |
| 10 |
Hb_003211_050 |
0.1696133072 |
- |
- |
PREDICTED: RNA-binding protein 24-B isoform X1 [Jatropha curcas] |
| 11 |
Hb_002099_050 |
0.1703945817 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_006816_440 |
0.1781747583 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 13 |
Hb_000300_400 |
0.1782723092 |
- |
- |
hypothetical protein PHAVU_001G114100g [Phaseolus vulgaris] |
| 14 |
Hb_002235_230 |
0.1797029331 |
- |
- |
latex abundant protein 1 [Hevea brasiliensis] |
| 15 |
Hb_000603_210 |
0.1797945051 |
- |
- |
hypothetical protein POPTR_0006s23660g [Populus trichocarpa] |
| 16 |
Hb_048937_030 |
0.1847357783 |
- |
- |
hypothetical protein RCOM_0231790 [Ricinus communis] |
| 17 |
Hb_001741_090 |
0.1851112978 |
rubber biosynthesis |
Gene Name: SRPP1 |
RecName: Full=Small rubber particle protein; Short=SRPP; AltName: Full=22 kDa rubber particle protein; Short=22 kDa RPP; AltName: Full=27 kDa natural rubber allergen; AltName: Full=Latex allergen Hev b 3; AltName: Allergen=Hev b 3 [Hevea brasiliensis] |
| 18 |
Hb_002260_090 |
0.1852238827 |
- |
- |
PREDICTED: uncharacterized protein LOC105636025 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000803_050 |
0.1876134605 |
- |
- |
PREDICTED: uncharacterized protein LOC105648296 [Jatropha curcas] |
| 20 |
Hb_078633_010 |
0.1886283229 |
- |
- |
PREDICTED: RNA-binding protein 24-B isoform X1 [Jatropha curcas] |