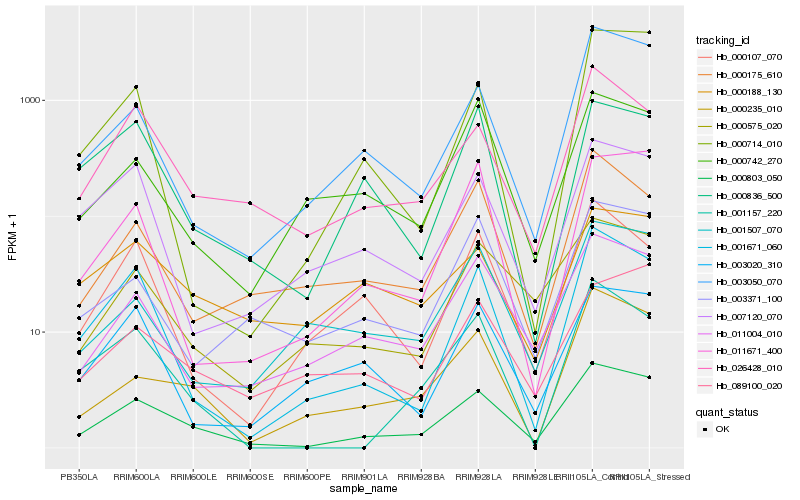
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000803_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648296 [Jatropha curcas] |
| 2 |
Hb_026428_010 |
0.1475656266 |
- |
- |
copper transport protein ATOX1 [Hevea brasiliensis] |
| 3 |
Hb_000235_010 |
0.1496836742 |
- |
- |
PREDICTED: serine carboxypeptidase-like 18 [Jatropha curcas] |
| 4 |
Hb_003050_070 |
0.1504528753 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_011004_010 |
0.1507550836 |
- |
- |
Mitochondrial carnitine/acylcarnitine carrier protein, putative [Ricinus communis] |
| 6 |
Hb_001671_060 |
0.1563503657 |
- |
- |
fad NAD binding oxidoreductases, putative [Ricinus communis] |
| 7 |
Hb_000575_020 |
0.1588316972 |
- |
- |
PREDICTED: copper-transporting ATPase PAA1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_003371_100 |
0.1731427498 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 9 |
Hb_011671_400 |
0.1746269099 |
- |
- |
PREDICTED: cyclin-U4-1 [Jatropha curcas] |
| 10 |
Hb_000175_610 |
0.1748508193 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 8 isoform X3 [Jatropha curcas] |
| 11 |
Hb_001157_220 |
0.1749172907 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_007120_070 |
0.1780914904 |
- |
- |
PREDICTED: caffeoylshikimate esterase-like [Jatropha curcas] |
| 13 |
Hb_001507_070 |
0.1788807457 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_089100_020 |
0.1797154425 |
- |
- |
PREDICTED: cyclin-A1-1 [Jatropha curcas] |
| 15 |
Hb_000188_130 |
0.1807545153 |
- |
- |
PREDICTED: uncharacterized protein LOC102628744 [Citrus sinensis] |
| 16 |
Hb_000836_500 |
0.185007512 |
- |
- |
Late embryogenesis abundant protein Lea14-A, putative [Ricinus communis] |
| 17 |
Hb_000742_270 |
0.1856356459 |
- |
- |
PREDICTED: fatty acid 2-hydroxylase 1-like [Jatropha curcas] |
| 18 |
Hb_003020_310 |
0.1857856633 |
- |
- |
glycosyltransferase, putative [Ricinus communis] |
| 19 |
Hb_000714_010 |
0.1865045198 |
- |
- |
PREDICTED: salt stress-induced hydrophobic peptide ESI3 [Vitis vinifera] |
| 20 |
Hb_000107_070 |
0.1867589352 |
- |
- |
PREDICTED: uncharacterized protein LOC105641007 [Jatropha curcas] |