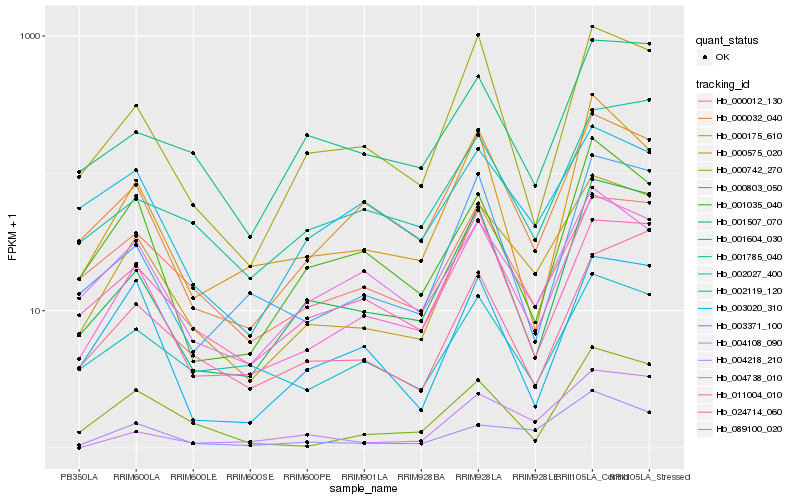
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000575_020 |
0.0 |
- |
- |
PREDICTED: copper-transporting ATPase PAA1, chloroplastic [Jatropha curcas] |
| 2 |
Hb_011004_010 |
0.0965184967 |
- |
- |
Mitochondrial carnitine/acylcarnitine carrier protein, putative [Ricinus communis] |
| 3 |
Hb_004108_090 |
0.1187717502 |
- |
- |
PREDICTED: uncharacterized protein LOC105650231 [Jatropha curcas] |
| 4 |
Hb_000032_040 |
0.1416851244 |
- |
- |
PREDICTED: mitochondrial adenine nucleotide transporter ADNT1 [Jatropha curcas] |
| 5 |
Hb_001507_070 |
0.1440475485 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000742_270 |
0.1521447943 |
- |
- |
PREDICTED: fatty acid 2-hydroxylase 1-like [Jatropha curcas] |
| 7 |
Hb_000012_130 |
0.1579697156 |
- |
- |
PREDICTED: uncharacterized protein LOC100807540 isoform X2 [Glycine max] |
| 8 |
Hb_000803_050 |
0.1588316972 |
- |
- |
PREDICTED: uncharacterized protein LOC105648296 [Jatropha curcas] |
| 9 |
Hb_003371_100 |
0.1605471292 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 10 |
Hb_004738_010 |
0.1619637748 |
- |
- |
PREDICTED: N-acetyl-D-glucosamine kinase-like [Jatropha curcas] |
| 11 |
Hb_001785_040 |
0.165457727 |
- |
- |
cytochrome b5 isoform Cb5-A [Vernicia fordii] |
| 12 |
Hb_001604_030 |
0.1668969582 |
- |
- |
ubiquitin-conjugating enzyme E2 35-like [Cucumis sativus] |
| 13 |
Hb_003020_310 |
0.1669925777 |
- |
- |
glycosyltransferase, putative [Ricinus communis] |
| 14 |
Hb_089100_020 |
0.1696141171 |
- |
- |
PREDICTED: cyclin-A1-1 [Jatropha curcas] |
| 15 |
Hb_002027_400 |
0.1696872246 |
- |
- |
PREDICTED: auxin response factor 23-like [Jatropha curcas] |
| 16 |
Hb_001035_040 |
0.1705627336 |
- |
- |
unnamed protein product [Coffea canephora] |
| 17 |
Hb_000175_610 |
0.1732066998 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 8 isoform X3 [Jatropha curcas] |
| 18 |
Hb_002119_120 |
0.1732494102 |
- |
- |
PREDICTED: anthranilate synthase beta subunit 1, chloroplastic-like [Jatropha curcas] |
| 19 |
Hb_004218_210 |
0.1732845047 |
transcription factor |
TF Family: Orphans |
hypothetical protein JCGZ_24525 [Jatropha curcas] |
| 20 |
Hb_024714_060 |
0.1759093351 |
- |
- |
PREDICTED: thiamine pyrophosphokinase 1 isoform X1 [Jatropha curcas] |