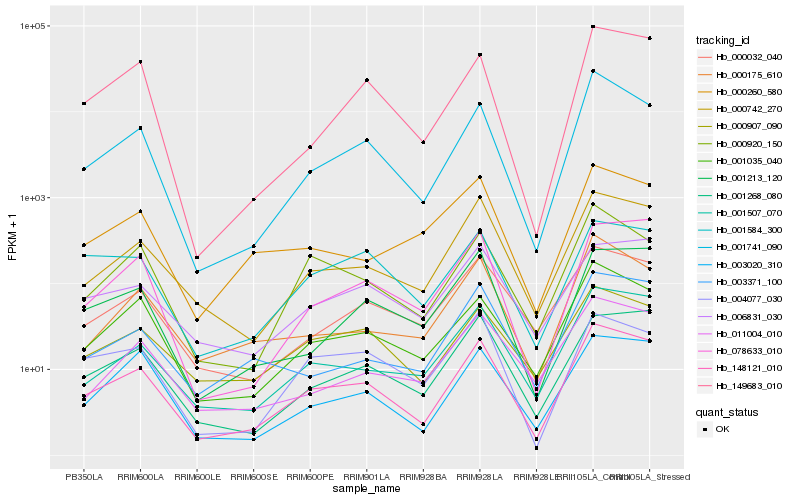
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_148121_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000742_270 |
0.0960910647 |
- |
- |
PREDICTED: fatty acid 2-hydroxylase 1-like [Jatropha curcas] |
| 3 |
Hb_001741_090 |
0.1044232932 |
rubber biosynthesis |
Gene Name: SRPP1 |
RecName: Full=Small rubber particle protein; Short=SRPP; AltName: Full=22 kDa rubber particle protein; Short=22 kDa RPP; AltName: Full=27 kDa natural rubber allergen; AltName: Full=Latex allergen Hev b 3; AltName: Allergen=Hev b 3 [Hevea brasiliensis] |
| 4 |
Hb_001507_070 |
0.1097352397 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001035_040 |
0.1174413363 |
- |
- |
unnamed protein product [Coffea canephora] |
| 6 |
Hb_001268_080 |
0.1181019649 |
- |
- |
catalytic, putative [Ricinus communis] |
| 7 |
Hb_000032_040 |
0.1207963564 |
- |
- |
PREDICTED: mitochondrial adenine nucleotide transporter ADNT1 [Jatropha curcas] |
| 8 |
Hb_003020_310 |
0.1245836508 |
- |
- |
glycosyltransferase, putative [Ricinus communis] |
| 9 |
Hb_000907_090 |
0.1247700394 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 10 |
Hb_078633_010 |
0.1266867369 |
- |
- |
PREDICTED: RNA-binding protein 24-B isoform X1 [Jatropha curcas] |
| 11 |
Hb_000920_150 |
0.1292202234 |
- |
- |
annexin [Manihot esculenta] |
| 12 |
Hb_001213_120 |
0.1321466015 |
- |
- |
PREDICTED: immediate early response 3-interacting protein 1-like [Brassica rapa] |
| 13 |
Hb_000175_610 |
0.1342513922 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 8 isoform X3 [Jatropha curcas] |
| 14 |
Hb_011004_010 |
0.1349792396 |
- |
- |
Mitochondrial carnitine/acylcarnitine carrier protein, putative [Ricinus communis] |
| 15 |
Hb_003371_100 |
0.1350487147 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 16 |
Hb_006831_030 |
0.1371383307 |
- |
- |
PREDICTED: ras-related protein RABB1c [Fragaria vesca subsp. vesca] |
| 17 |
Hb_149683_010 |
0.1371516803 |
rubber biosynthesis |
Gene Name: REF2 |
RecName: Full=Rubber elongation factor protein; Short=REF; AltName: Allergen=Hev b 1 [Hevea brasiliensis] |
| 18 |
Hb_000260_580 |
0.1401437041 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001584_300 |
0.1433504206 |
- |
- |
PREDICTED: pyroglutamyl-peptidase 1-like protein [Jatropha curcas] |
| 20 |
Hb_004077_030 |
0.143548191 |
- |
- |
hypothetical protein EUGRSUZ_K01571 [Eucalyptus grandis] |