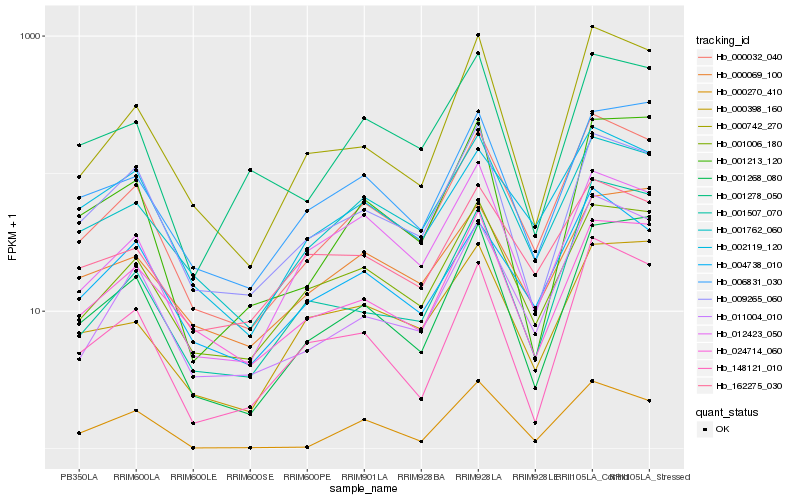
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000032_040 |
0.0 |
- |
- |
PREDICTED: mitochondrial adenine nucleotide transporter ADNT1 [Jatropha curcas] |
| 2 |
Hb_011004_010 |
0.083708911 |
- |
- |
Mitochondrial carnitine/acylcarnitine carrier protein, putative [Ricinus communis] |
| 3 |
Hb_001762_060 |
0.0897046242 |
- |
- |
pantothenate kinase, putative [Ricinus communis] |
| 4 |
Hb_001006_180 |
0.0945851877 |
- |
- |
PREDICTED: endonuclease V-like [Jatropha curcas] |
| 5 |
Hb_000742_270 |
0.0986131715 |
- |
- |
PREDICTED: fatty acid 2-hydroxylase 1-like [Jatropha curcas] |
| 6 |
Hb_004738_010 |
0.1032605774 |
- |
- |
PREDICTED: N-acetyl-D-glucosamine kinase-like [Jatropha curcas] |
| 7 |
Hb_002119_120 |
0.1127771829 |
- |
- |
PREDICTED: anthranilate synthase beta subunit 1, chloroplastic-like [Jatropha curcas] |
| 8 |
Hb_001268_080 |
0.1134069912 |
- |
- |
catalytic, putative [Ricinus communis] |
| 9 |
Hb_006831_030 |
0.1139235599 |
- |
- |
PREDICTED: ras-related protein RABB1c [Fragaria vesca subsp. vesca] |
| 10 |
Hb_024714_060 |
0.113927307 |
- |
- |
PREDICTED: thiamine pyrophosphokinase 1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_148121_010 |
0.1207963564 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_009265_060 |
0.1215793497 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme [Jatropha curcas] |
| 13 |
Hb_001507_070 |
0.121584427 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000069_100 |
0.1236251509 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_012423_050 |
0.1262398937 |
- |
- |
PREDICTED: transmembrane protein 64 [Jatropha curcas] |
| 16 |
Hb_000398_160 |
0.1265077101 |
- |
- |
GDP-mannose transporter, putative [Ricinus communis] |
| 17 |
Hb_001278_050 |
0.127152976 |
- |
- |
PREDICTED: uncharacterized protein LOC105636924 [Jatropha curcas] |
| 18 |
Hb_162275_030 |
0.1280545987 |
- |
- |
PREDICTED: uncharacterized protein LOC105646638 [Jatropha curcas] |
| 19 |
Hb_001213_120 |
0.1300924787 |
- |
- |
PREDICTED: immediate early response 3-interacting protein 1-like [Brassica rapa] |
| 20 |
Hb_000270_410 |
0.1326375021 |
- |
- |
- |