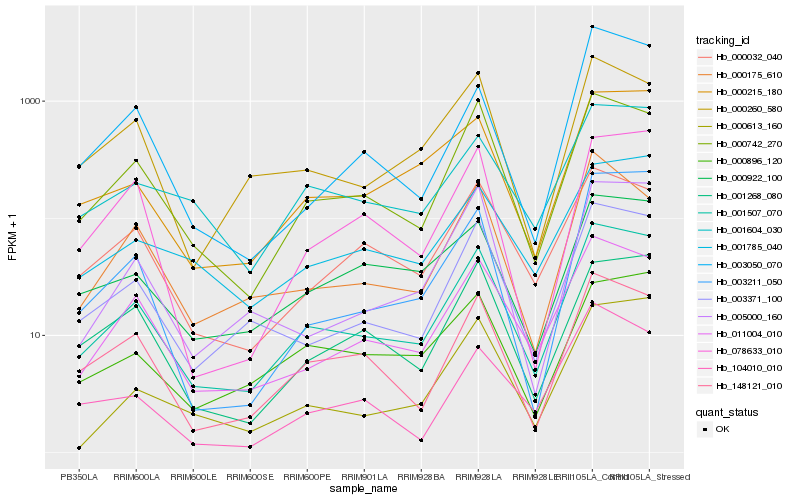
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001507_070 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000742_270 |
0.0854728121 |
- |
- |
PREDICTED: fatty acid 2-hydroxylase 1-like [Jatropha curcas] |
| 3 |
Hb_011004_010 |
0.092217753 |
- |
- |
Mitochondrial carnitine/acylcarnitine carrier protein, putative [Ricinus communis] |
| 4 |
Hb_003371_100 |
0.0981200027 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 5 |
Hb_148121_010 |
0.1097352397 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000215_180 |
0.1109294703 |
- |
- |
hypothetical protein 17 [Hevea brasiliensis] |
| 7 |
Hb_000032_040 |
0.121584427 |
- |
- |
PREDICTED: mitochondrial adenine nucleotide transporter ADNT1 [Jatropha curcas] |
| 8 |
Hb_001268_080 |
0.1234589166 |
- |
- |
catalytic, putative [Ricinus communis] |
| 9 |
Hb_001604_030 |
0.1249404115 |
- |
- |
ubiquitin-conjugating enzyme E2 35-like [Cucumis sativus] |
| 10 |
Hb_000896_120 |
0.1288141119 |
- |
- |
PREDICTED: acyl-protein thioesterase 2-like [Populus euphratica] |
| 11 |
Hb_000922_100 |
0.1290560229 |
- |
- |
PREDICTED: putative exosome complex component rrp40 [Jatropha curcas] |
| 12 |
Hb_000175_610 |
0.1299754946 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 8 isoform X3 [Jatropha curcas] |
| 13 |
Hb_005000_160 |
0.1304167184 |
- |
- |
PREDICTED: uncharacterized protein LOC105637882 [Jatropha curcas] |
| 14 |
Hb_000260_580 |
0.1304473222 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001785_040 |
0.1313638527 |
- |
- |
cytochrome b5 isoform Cb5-A [Vernicia fordii] |
| 16 |
Hb_078633_010 |
0.1346250389 |
- |
- |
PREDICTED: RNA-binding protein 24-B isoform X1 [Jatropha curcas] |
| 17 |
Hb_000613_160 |
0.135367675 |
- |
- |
coated vesicle membrane protein, putative [Ricinus communis] |
| 18 |
Hb_003211_050 |
0.1360021767 |
- |
- |
PREDICTED: RNA-binding protein 24-B isoform X1 [Jatropha curcas] |
| 19 |
Hb_003050_070 |
0.1413118026 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_104010_010 |
0.1413279531 |
- |
- |
hypothetical protein JCGZ_26423 [Jatropha curcas] |