| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
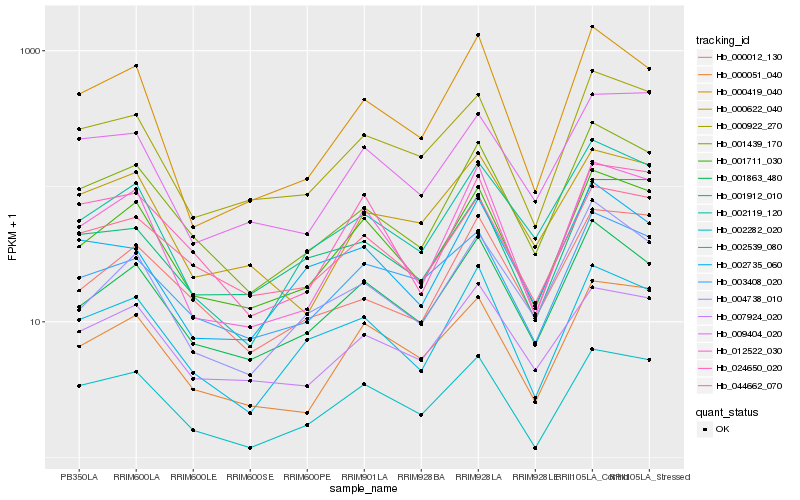
Hb_001439_170 |
0.0 |
- |
- |
PREDICTED: pectinesterase 31-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_000012_130 |
0.0849948205 |
- |
- |
PREDICTED: uncharacterized protein LOC100807540 isoform X2 [Glycine max] |
| 3 |
Hb_001711_030 |
0.0850174285 |
- |
- |
PREDICTED: uncharacterized protein LOC105642409 [Jatropha curcas] |
| 4 |
Hb_001863_480 |
0.0854812216 |
- |
- |
PREDICTED: uncharacterized protein LOC105630612 [Jatropha curcas] |
| 5 |
Hb_002119_120 |
0.088757207 |
- |
- |
PREDICTED: anthranilate synthase beta subunit 1, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_044662_070 |
0.0915378939 |
- |
- |
PREDICTED: hsp70-Hsp90 organizing protein 3 [Jatropha curcas] |
| 7 |
Hb_002539_080 |
0.0956810473 |
- |
- |
PREDICTED: triose phosphate/phosphate translocator, chloroplastic-like [Jatropha curcas] |
| 8 |
Hb_000922_270 |
0.097702882 |
- |
- |
iron-sulfur cluster scaffold protein [Hevea brasiliensis] |
| 9 |
Hb_007924_020 |
0.0984147647 |
- |
- |
PREDICTED: chaperonin CPN60-like 2, mitochondrial isoform X2 [Jatropha curcas] |
| 10 |
Hb_004738_010 |
0.099568417 |
- |
- |
PREDICTED: N-acetyl-D-glucosamine kinase-like [Jatropha curcas] |
| 11 |
Hb_003408_020 |
0.1086315434 |
- |
- |
hypothetical protein RCOM_0453040 [Ricinus communis] |
| 12 |
Hb_012522_030 |
0.1088285596 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_000419_040 |
0.1099504563 |
- |
- |
chorismate synthase [Hevea brasiliensis] |
| 14 |
Hb_000622_040 |
0.1121026582 |
- |
- |
PREDICTED: uncharacterized protein LOC104596457 isoform X1 [Nelumbo nucifera] |
| 15 |
Hb_002282_020 |
0.1128738727 |
- |
- |
PREDICTED: uncharacterized protein LOC105629828 [Jatropha curcas] |
| 16 |
Hb_000051_040 |
0.1142257589 |
- |
- |
PREDICTED: uncharacterized protein LOC102597788 [Solanum tuberosum] |
| 17 |
Hb_002735_060 |
0.1162518193 |
- |
- |
PREDICTED: desumoylating isopeptidase 1 [Jatropha curcas] |
| 18 |
Hb_024650_020 |
0.1168191226 |
- |
- |
PREDICTED: uncharacterized protein LOC105646626 [Jatropha curcas] |
| 19 |
Hb_001912_010 |
0.1191821293 |
- |
- |
PREDICTED: uncharacterized protein LOC105636600 [Jatropha curcas] |
| 20 |
Hb_009404_020 |
0.1192694789 |
- |
- |
conserved hypothetical protein [Ricinus communis] |