| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
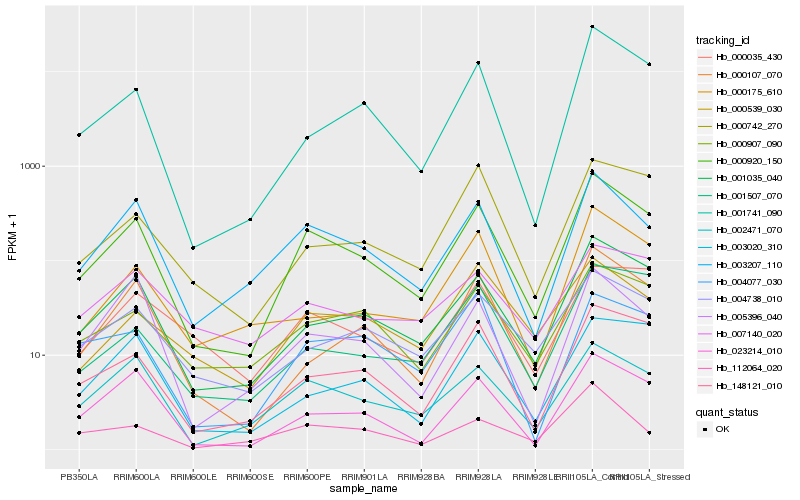
Hb_000920_150 |
0.0 |
- |
- |
annexin [Manihot esculenta] |
| 2 |
Hb_001035_040 |
0.1101688128 |
- |
- |
unnamed protein product [Coffea canephora] |
| 3 |
Hb_003207_110 |
0.1150199289 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A synthase |
hydroxymethylglutaryl coenzyme A synthase [Hevea brasiliensis] |
| 4 |
Hb_148121_010 |
0.1292202234 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_023214_010 |
0.1357522018 |
- |
- |
aberrant lateral root formation 1 family protein [Populus trichocarpa] |
| 6 |
Hb_002471_070 |
0.1362342412 |
- |
- |
Tartrate-resistant acid phosphatase type 5 precursor, putative [Ricinus communis] |
| 7 |
Hb_001741_090 |
0.142760029 |
rubber biosynthesis |
Gene Name: SRPP1 |
RecName: Full=Small rubber particle protein; Short=SRPP; AltName: Full=22 kDa rubber particle protein; Short=22 kDa RPP; AltName: Full=27 kDa natural rubber allergen; AltName: Full=Latex allergen Hev b 3; AltName: Allergen=Hev b 3 [Hevea brasiliensis] |
| 8 |
Hb_000907_090 |
0.1503301309 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 9 |
Hb_000107_070 |
0.1518990749 |
- |
- |
PREDICTED: uncharacterized protein LOC105641007 [Jatropha curcas] |
| 10 |
Hb_004738_010 |
0.1595671932 |
- |
- |
PREDICTED: N-acetyl-D-glucosamine kinase-like [Jatropha curcas] |
| 11 |
Hb_000175_610 |
0.1666950961 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 8 isoform X3 [Jatropha curcas] |
| 12 |
Hb_000742_270 |
0.1691271671 |
- |
- |
PREDICTED: fatty acid 2-hydroxylase 1-like [Jatropha curcas] |
| 13 |
Hb_000539_030 |
0.1746132711 |
rubber biosynthesis |
Gene Name: Mevalonate diphosphate decarboxylase |
diphosphomevelonate decarboxylase [Hevea brasiliensis] |
| 14 |
Hb_003020_310 |
0.1747750272 |
- |
- |
glycosyltransferase, putative [Ricinus communis] |
| 15 |
Hb_001507_070 |
0.1778327558 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000035_430 |
0.1792816809 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2-like [Jatropha curcas] |
| 17 |
Hb_004077_030 |
0.1798661261 |
- |
- |
hypothetical protein EUGRSUZ_K01571 [Eucalyptus grandis] |
| 18 |
Hb_112064_020 |
0.1824860165 |
- |
- |
PREDICTED: cytochrome b561 and DOMON domain-containing protein At5g47530-like [Jatropha curcas] |
| 19 |
Hb_005396_040 |
0.1824999052 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 20 |
Hb_007140_020 |
0.1826081972 |
- |
- |
hypothetical protein CICLE_v10008419mg [Citrus clementina] |