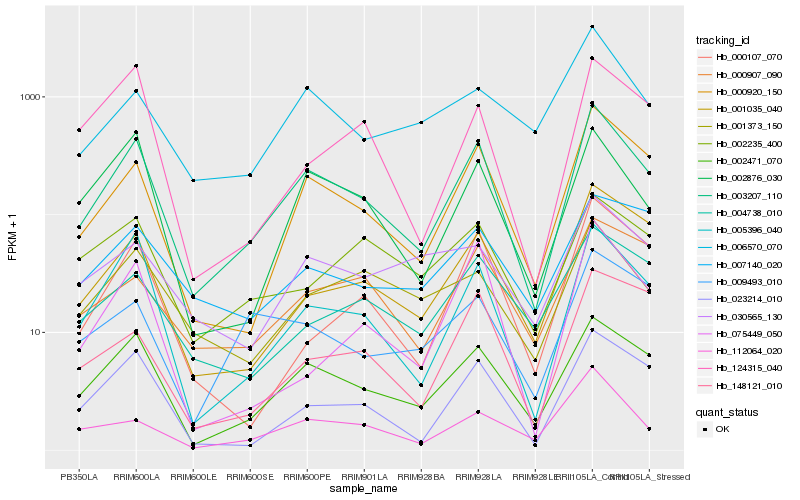
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003207_110 |
0.0 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A synthase |
hydroxymethylglutaryl coenzyme A synthase [Hevea brasiliensis] |
| 2 |
Hb_002471_070 |
0.0982031278 |
- |
- |
Tartrate-resistant acid phosphatase type 5 precursor, putative [Ricinus communis] |
| 3 |
Hb_000920_150 |
0.1150199289 |
- |
- |
annexin [Manihot esculenta] |
| 4 |
Hb_005396_040 |
0.123735742 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 5 |
Hb_023214_010 |
0.1443858333 |
- |
- |
aberrant lateral root formation 1 family protein [Populus trichocarpa] |
| 6 |
Hb_002876_030 |
0.1511411453 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 4 [Jatropha curcas] |
| 7 |
Hb_001035_040 |
0.1561616011 |
- |
- |
unnamed protein product [Coffea canephora] |
| 8 |
Hb_112064_020 |
0.1680721817 |
- |
- |
PREDICTED: cytochrome b561 and DOMON domain-containing protein At5g47530-like [Jatropha curcas] |
| 9 |
Hb_000907_090 |
0.1774681868 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 10 |
Hb_001373_150 |
0.1789053445 |
- |
- |
PREDICTED: uncharacterized protein LOC105628778 [Jatropha curcas] |
| 11 |
Hb_004738_010 |
0.1827120194 |
- |
- |
PREDICTED: N-acetyl-D-glucosamine kinase-like [Jatropha curcas] |
| 12 |
Hb_006570_070 |
0.1840032715 |
- |
- |
cyclophilin [Hevea brasiliensis] |
| 13 |
Hb_000107_070 |
0.1863702546 |
- |
- |
PREDICTED: uncharacterized protein LOC105641007 [Jatropha curcas] |
| 14 |
Hb_124315_040 |
0.1885480665 |
- |
- |
hypothetical protein 19 [Hevea brasiliensis] |
| 15 |
Hb_002235_400 |
0.1890788328 |
- |
- |
PREDICTED: uncharacterized protein At5g19025-like isoform X1 [Populus euphratica] |
| 16 |
Hb_148121_010 |
0.190385528 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_009493_010 |
0.1910519515 |
- |
- |
Minor allergen Alt a, putative [Ricinus communis] |
| 18 |
Hb_007140_020 |
0.19120278 |
- |
- |
hypothetical protein CICLE_v10008419mg [Citrus clementina] |
| 19 |
Hb_075449_050 |
0.1950435508 |
- |
- |
PREDICTED: premnaspirodiene oxygenase-like [Jatropha curcas] |
| 20 |
Hb_030565_130 |
0.1953792071 |
- |
- |
hypothetical protein JCGZ_01083 [Jatropha curcas] |