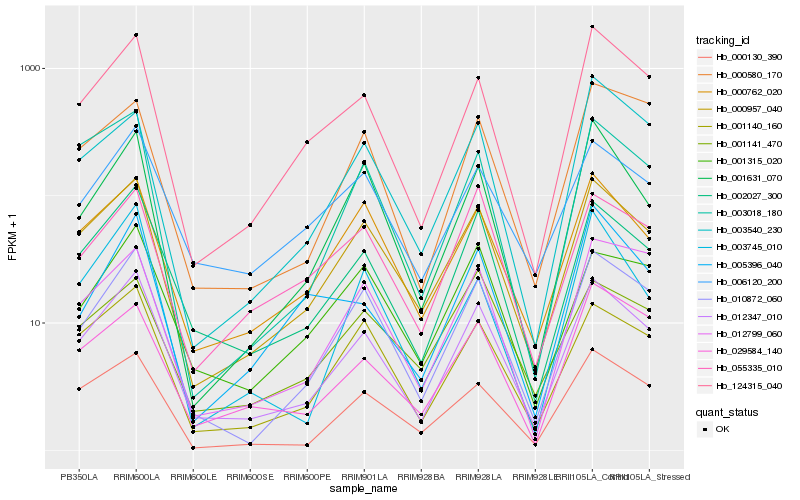
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012347_010 |
0.0 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_03155 [Jatropha curcas] |
| 2 |
Hb_002027_300 |
0.0605732258 |
- |
- |
PREDICTED: rho GTPase-activating protein 2 [Jatropha curcas] |
| 3 |
Hb_000957_040 |
0.0764853831 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RNF144A-B [Jatropha curcas] |
| 4 |
Hb_010872_060 |
0.0872591572 |
transcription factor |
TF Family: MYB |
Duplicated homeodomain-like superfamily protein, putative [Theobroma cacao] |
| 5 |
Hb_124315_040 |
0.0969397406 |
- |
- |
hypothetical protein 19 [Hevea brasiliensis] |
| 6 |
Hb_001140_160 |
0.100497156 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 7 |
Hb_000130_390 |
0.1069461304 |
- |
- |
PREDICTED: glutamate receptor 2.8-like [Pyrus x bretschneideri] |
| 8 |
Hb_000762_020 |
0.1108264387 |
- |
- |
PREDICTED: uncharacterized protein LOC105641515 [Jatropha curcas] |
| 9 |
Hb_006120_200 |
0.1220278914 |
- |
- |
PREDICTED: uncharacterized protein LOC103333282 [Prunus mume] |
| 10 |
Hb_001631_070 |
0.1242469525 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 11 |
Hb_003018_180 |
0.1269559515 |
- |
- |
PREDICTED: WEB family protein At3g51220-like [Jatropha curcas] |
| 12 |
Hb_029584_140 |
0.1306349461 |
- |
- |
senescence-associated family protein [Populus trichocarpa] |
| 13 |
Hb_001315_020 |
0.1310023361 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 14 |
Hb_012799_060 |
0.1375021178 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 15 |
Hb_055335_010 |
0.1385428555 |
- |
- |
PREDICTED: tetraspanin-2 [Jatropha curcas] |
| 16 |
Hb_000580_170 |
0.1404546013 |
- |
- |
PREDICTED: arginase 1, mitochondrial [Jatropha curcas] |
| 17 |
Hb_003745_010 |
0.1438689456 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_03155 [Jatropha curcas] |
| 18 |
Hb_003540_230 |
0.1455047032 |
- |
- |
PREDICTED: sphingosine kinase 1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001141_470 |
0.1516253048 |
- |
- |
PREDICTED: uncharacterized protein LOC101254423 [Solanum lycopersicum] |
| 20 |
Hb_005396_040 |
0.1523745018 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |