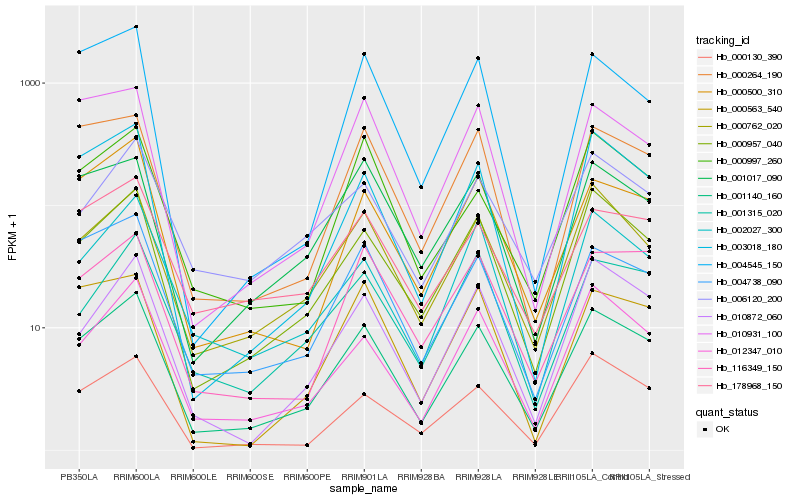
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001140_160 |
0.0 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 2 |
Hb_000957_040 |
0.0759797198 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RNF144A-B [Jatropha curcas] |
| 3 |
Hb_000762_020 |
0.0903548605 |
- |
- |
PREDICTED: uncharacterized protein LOC105641515 [Jatropha curcas] |
| 4 |
Hb_004738_090 |
0.097409231 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_003018_180 |
0.0994981364 |
- |
- |
PREDICTED: WEB family protein At3g51220-like [Jatropha curcas] |
| 6 |
Hb_012347_010 |
0.100497156 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_03155 [Jatropha curcas] |
| 7 |
Hb_000500_310 |
0.1039149122 |
- |
- |
- |
| 8 |
Hb_010872_060 |
0.1081724875 |
transcription factor |
TF Family: MYB |
Duplicated homeodomain-like superfamily protein, putative [Theobroma cacao] |
| 9 |
Hb_002027_300 |
0.1107788038 |
- |
- |
PREDICTED: rho GTPase-activating protein 2 [Jatropha curcas] |
| 10 |
Hb_000264_190 |
0.1117680253 |
- |
- |
sphingolipid delta 4 desaturase/C-4 hydroxylase protein des2, putative [Ricinus communis] |
| 11 |
Hb_000130_390 |
0.1130163931 |
- |
- |
PREDICTED: glutamate receptor 2.8-like [Pyrus x bretschneideri] |
| 12 |
Hb_001017_090 |
0.1164921313 |
- |
- |
- |
| 13 |
Hb_116349_150 |
0.1175556223 |
- |
- |
PREDICTED: microtubule-associated protein TORTIFOLIA1 [Jatropha curcas] |
| 14 |
Hb_001315_020 |
0.1209695274 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 15 |
Hb_178968_150 |
0.1209809168 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 16 |
Hb_000997_260 |
0.1222996895 |
- |
- |
PREDICTED: rRNA-processing protein EFG1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_010931_100 |
0.1231938681 |
- |
- |
PREDICTED: uncharacterized protein LOC105638669 [Jatropha curcas] |
| 18 |
Hb_006120_200 |
0.1262834445 |
- |
- |
PREDICTED: uncharacterized protein LOC103333282 [Prunus mume] |
| 19 |
Hb_000563_540 |
0.1302516328 |
- |
- |
PREDICTED: Fanconi anemia group I protein [Jatropha curcas] |
| 20 |
Hb_004545_150 |
0.1305715501 |
- |
- |
PREDICTED: paramyosin [Jatropha curcas] |