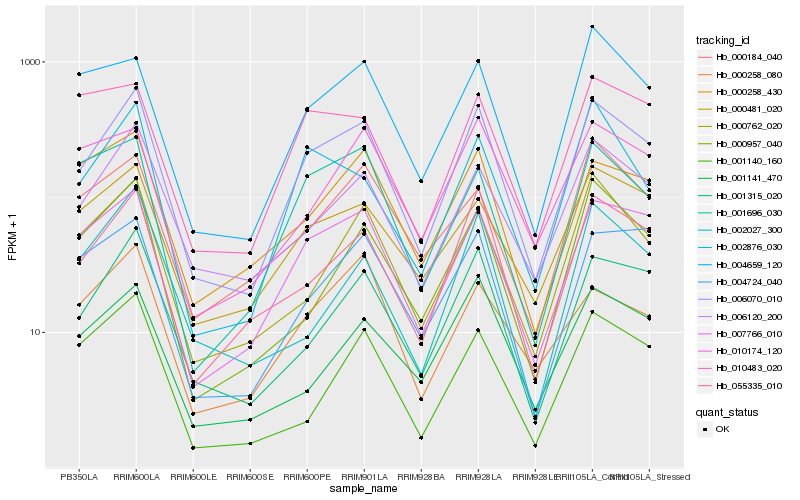
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006070_010 |
0.0 |
rubber biosynthesis |
Gene Name: CPT2 |
cis-prenyltransferase 3 [Hevea brasiliensis] |
| 2 |
Hb_007766_010 |
0.0929586474 |
- |
- |
hypothetical protein JCGZ_06718 [Jatropha curcas] |
| 3 |
Hb_055335_010 |
0.1080324658 |
- |
- |
PREDICTED: tetraspanin-2 [Jatropha curcas] |
| 4 |
Hb_001315_020 |
0.1105472645 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 5 |
Hb_006120_200 |
0.1138925216 |
- |
- |
PREDICTED: uncharacterized protein LOC103333282 [Prunus mume] |
| 6 |
Hb_000481_020 |
0.1279105815 |
- |
- |
PREDICTED: shikimate kinase 1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000762_020 |
0.1322006034 |
- |
- |
PREDICTED: uncharacterized protein LOC105641515 [Jatropha curcas] |
| 8 |
Hb_004659_120 |
0.1402724535 |
rubber biosynthesis |
Gene Name: CPTL |
PREDICTED: dehydrodolichyl diphosphate syntase complex subunit NUS1 isoform X3 [Jatropha curcas] |
| 9 |
Hb_000957_040 |
0.1419292982 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RNF144A-B [Jatropha curcas] |
| 10 |
Hb_004724_040 |
0.1420492454 |
- |
- |
PREDICTED: uncharacterized protein LOC105640153 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002876_030 |
0.1431250954 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 4 [Jatropha curcas] |
| 12 |
Hb_000258_430 |
0.1433582915 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 13 |
Hb_001696_030 |
0.1447408333 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 14 |
Hb_001141_470 |
0.1447823474 |
- |
- |
PREDICTED: uncharacterized protein LOC101254423 [Solanum lycopersicum] |
| 15 |
Hb_000184_040 |
0.1494325698 |
rubber biosynthesis |
Gene Name: Acetyl-CoA acetyltransferase |
PREDICTED: acetyl-CoA acetyltransferase, cytosolic 1-like isoform X2 [Nelumbo nucifera] |
| 16 |
Hb_010174_120 |
0.1497584852 |
- |
- |
PREDICTED: very-long-chain 3-oxoacyl-CoA reductase 1-like [Gossypium raimondii] |
| 17 |
Hb_001140_160 |
0.1521836122 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 18 |
Hb_000258_080 |
0.1534762193 |
- |
- |
Brassinosteroid LRR receptor kinase precursor, putative [Ricinus communis] |
| 19 |
Hb_010483_020 |
0.1541351187 |
- |
- |
latex allergen [Hevea brasiliensis] |
| 20 |
Hb_002027_300 |
0.1542850806 |
- |
- |
PREDICTED: rho GTPase-activating protein 2 [Jatropha curcas] |