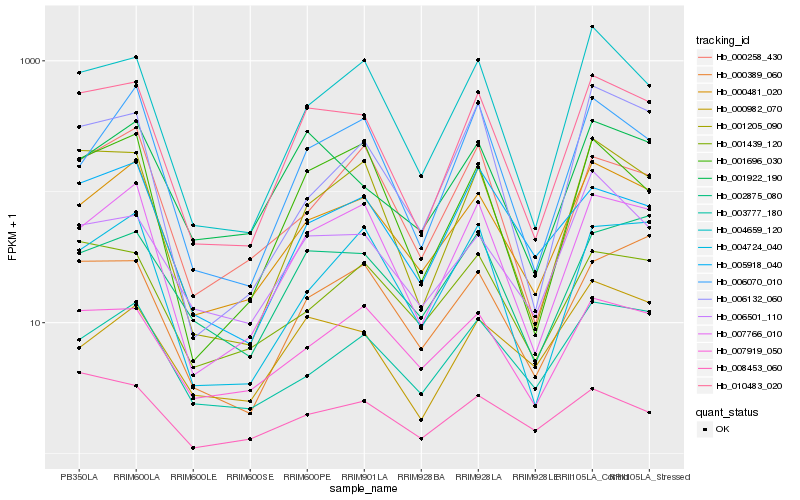
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010483_020 |
0.0 |
- |
- |
latex allergen [Hevea brasiliensis] |
| 2 |
Hb_001205_090 |
0.1021491872 |
- |
- |
PREDICTED: random slug protein 5 [Jatropha curcas] |
| 3 |
Hb_007766_010 |
0.1037792481 |
- |
- |
hypothetical protein JCGZ_06718 [Jatropha curcas] |
| 4 |
Hb_000481_020 |
0.1148487997 |
- |
- |
PREDICTED: shikimate kinase 1, chloroplastic [Jatropha curcas] |
| 5 |
Hb_004659_120 |
0.1220929968 |
rubber biosynthesis |
Gene Name: CPTL |
PREDICTED: dehydrodolichyl diphosphate syntase complex subunit NUS1 isoform X3 [Jatropha curcas] |
| 6 |
Hb_001696_030 |
0.1275878663 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 7 |
Hb_001922_190 |
0.1302247339 |
- |
- |
RecName: Full=Hevamine-A; Includes: RecName: Full=Chitinase; Includes: RecName: Full=Lysozyme; Flags: Precursor [Hevea brasiliensis] |
| 8 |
Hb_002875_080 |
0.1380945864 |
- |
- |
protein with unknown function [Ricinus communis] |
| 9 |
Hb_005918_040 |
0.1399940969 |
- |
- |
hypothetical protein POPTR_0003s04560g [Populus trichocarpa] |
| 10 |
Hb_007919_050 |
0.1419769512 |
- |
- |
PREDICTED: uncharacterized protein LOC105635142 isoform X2 [Jatropha curcas] |
| 11 |
Hb_006501_110 |
0.1458214569 |
- |
- |
PREDICTED: probable 2-oxoglutarate-dependent dioxygenase At3g49630 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001439_120 |
0.148985371 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_006070_010 |
0.1541351187 |
rubber biosynthesis |
Gene Name: CPT2 |
cis-prenyltransferase 3 [Hevea brasiliensis] |
| 14 |
Hb_000389_060 |
0.1542617915 |
- |
- |
PREDICTED: synaptotagmin-3 isoform X1 [Jatropha curcas] |
| 15 |
Hb_008453_060 |
0.1548026504 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_004724_040 |
0.1555345164 |
- |
- |
PREDICTED: uncharacterized protein LOC105640153 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003777_180 |
0.158138206 |
- |
- |
PREDICTED: sterol 3-beta-glucosyltransferase UGT80B1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_006132_060 |
0.1582360847 |
- |
- |
phospholipid/glycerol acyltransferase family protein [Populus trichocarpa] |
| 19 |
Hb_000982_070 |
0.1596038052 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 20 |
Hb_000258_430 |
0.1598511012 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |