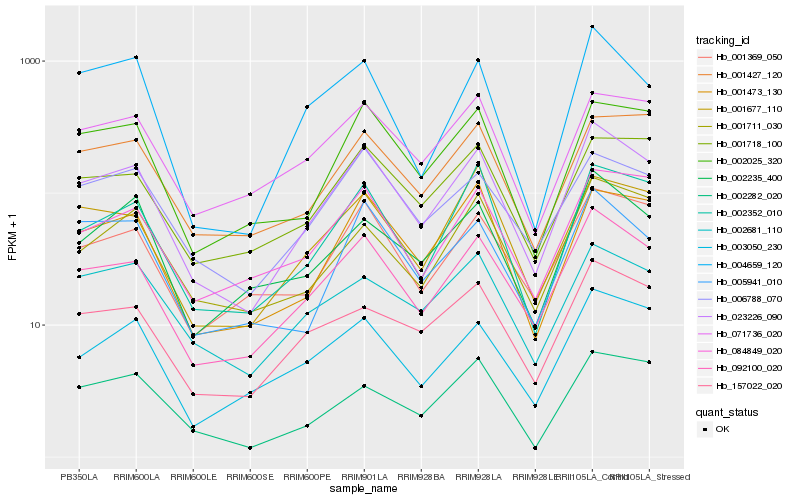
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_023226_090 |
0.0 |
- |
- |
PREDICTED: glycolipid transfer protein [Jatropha curcas] |
| 2 |
Hb_092100_020 |
0.0561333799 |
- |
- |
strictosidine synthase, putative [Ricinus communis] |
| 3 |
Hb_002352_010 |
0.0958999151 |
- |
- |
PREDICTED: rop guanine nucleotide exchange factor 1 [Jatropha curcas] |
| 4 |
Hb_001473_130 |
0.0980265347 |
- |
- |
PREDICTED: ubiquinone biosynthesis monooxygenase COQ6 [Jatropha curcas] |
| 5 |
Hb_002025_320 |
0.1022617269 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit I-like [Jatropha curcas] |
| 6 |
Hb_001369_050 |
0.102829356 |
- |
- |
hypothetical protein MIMGU_mgv1a0083172mg, partial [Erythranthe guttata] |
| 7 |
Hb_157022_020 |
0.1048961121 |
- |
- |
nicotinamide mononucleotide adenylyltransferase, putative [Ricinus communis] |
| 8 |
Hb_006788_070 |
0.107971641 |
- |
- |
2-methyl-6-geranylgeranylbenzoquinone methyltranferase [Hevea brasiliensis] |
| 9 |
Hb_003050_230 |
0.1080144316 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_005941_010 |
0.1083707124 |
- |
- |
PREDICTED: protein SAND [Jatropha curcas] |
| 11 |
Hb_084849_020 |
0.1085078374 |
- |
- |
ARF GTPase activator, putative [Ricinus communis] |
| 12 |
Hb_001677_110 |
0.1087643698 |
- |
- |
Protein YIPF1, putative [Ricinus communis] |
| 13 |
Hb_001718_100 |
0.1114816903 |
- |
- |
PREDICTED: casein kinase II subunit beta-like isoform X5 [Jatropha curcas] |
| 14 |
Hb_071736_020 |
0.1116457751 |
- |
- |
hypothetical protein POPTR_0006s10330g [Populus trichocarpa] |
| 15 |
Hb_004659_120 |
0.1124585489 |
rubber biosynthesis |
Gene Name: CPTL |
PREDICTED: dehydrodolichyl diphosphate syntase complex subunit NUS1 isoform X3 [Jatropha curcas] |
| 16 |
Hb_002235_400 |
0.1138858322 |
- |
- |
PREDICTED: uncharacterized protein At5g19025-like isoform X1 [Populus euphratica] |
| 17 |
Hb_001711_030 |
0.1144109858 |
- |
- |
PREDICTED: uncharacterized protein LOC105642409 [Jatropha curcas] |
| 18 |
Hb_001427_120 |
0.1144450724 |
- |
- |
PREDICTED: uncharacterized protein LOC105637787 [Jatropha curcas] |
| 19 |
Hb_002681_110 |
0.1149020647 |
- |
- |
PREDICTED: uncharacterized protein LOC105633759 [Jatropha curcas] |
| 20 |
Hb_002282_020 |
0.1157783298 |
- |
- |
PREDICTED: uncharacterized protein LOC105629828 [Jatropha curcas] |