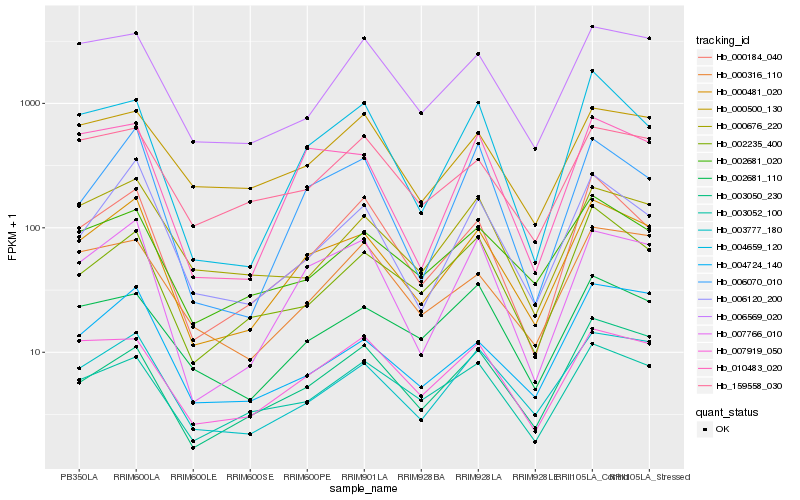
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000481_020 |
0.0 |
- |
- |
PREDICTED: shikimate kinase 1, chloroplastic [Jatropha curcas] |
| 2 |
Hb_003777_180 |
0.0863643434 |
- |
- |
PREDICTED: sterol 3-beta-glucosyltransferase UGT80B1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_007766_010 |
0.0940061198 |
- |
- |
hypothetical protein JCGZ_06718 [Jatropha curcas] |
| 4 |
Hb_000184_040 |
0.1061743382 |
rubber biosynthesis |
Gene Name: Acetyl-CoA acetyltransferase |
PREDICTED: acetyl-CoA acetyltransferase, cytosolic 1-like isoform X2 [Nelumbo nucifera] |
| 5 |
Hb_006120_200 |
0.1079742571 |
- |
- |
PREDICTED: uncharacterized protein LOC103333282 [Prunus mume] |
| 6 |
Hb_002681_020 |
0.1090666098 |
- |
- |
aldose 1-epimerase, putative [Ricinus communis] |
| 7 |
Hb_007919_050 |
0.1143102299 |
- |
- |
PREDICTED: uncharacterized protein LOC105635142 isoform X2 [Jatropha curcas] |
| 8 |
Hb_010483_020 |
0.1148487997 |
- |
- |
latex allergen [Hevea brasiliensis] |
| 9 |
Hb_003052_100 |
0.1172315963 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 10 |
Hb_002235_400 |
0.1204678257 |
- |
- |
PREDICTED: uncharacterized protein At5g19025-like isoform X1 [Populus euphratica] |
| 11 |
Hb_006569_020 |
0.1207778372 |
- |
- |
hypothetical protein POPTR_0010s22990g [Populus trichocarpa] |
| 12 |
Hb_003050_230 |
0.1210962507 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_159558_030 |
0.1246409093 |
- |
- |
ubiquitin-conjugating enzyme E2 variant 1D [Jatropha curcas] |
| 14 |
Hb_000676_220 |
0.1262366308 |
- |
- |
ADP-ribosylation factor, putative [Ricinus communis] |
| 15 |
Hb_004659_120 |
0.1264758884 |
rubber biosynthesis |
Gene Name: CPTL |
PREDICTED: dehydrodolichyl diphosphate syntase complex subunit NUS1 isoform X3 [Jatropha curcas] |
| 16 |
Hb_000316_110 |
0.1267916503 |
- |
- |
Dolichyl-phosphate beta-glucosyltransferase, putative [Ricinus communis] |
| 17 |
Hb_002681_110 |
0.1271299956 |
- |
- |
PREDICTED: uncharacterized protein LOC105633759 [Jatropha curcas] |
| 18 |
Hb_004724_140 |
0.1271776902 |
transcription factor |
TF Family: NF-YB |
PREDICTED: receptor protein kinase CLAVATA1 [Jatropha curcas] |
| 19 |
Hb_006070_010 |
0.1279105815 |
rubber biosynthesis |
Gene Name: CPT2 |
cis-prenyltransferase 3 [Hevea brasiliensis] |
| 20 |
Hb_000500_130 |
0.1280368426 |
- |
- |
small GTPase [Hevea brasiliensis] |