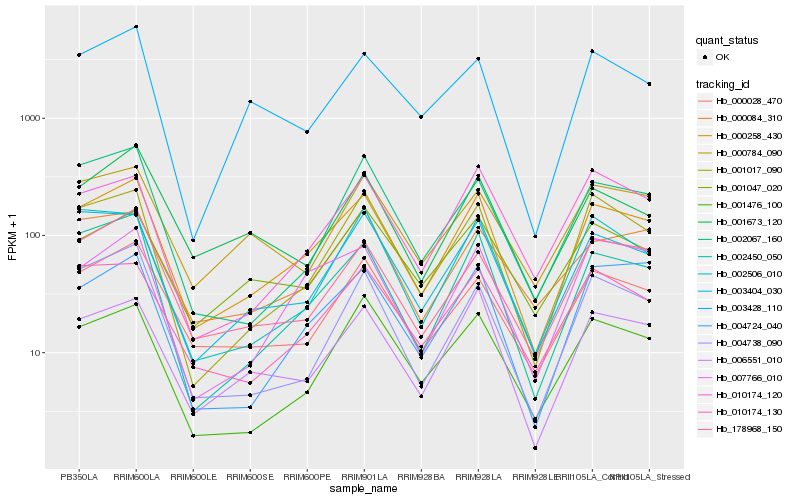
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000258_430 |
0.0 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 2 |
Hb_002450_050 |
0.0873634119 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like [Solanum lycopersicum] |
| 3 |
Hb_006551_010 |
0.0928034816 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 4 |
Hb_001017_090 |
0.0941748603 |
- |
- |
- |
| 5 |
Hb_000028_470 |
0.0973108802 |
transcription factor |
TF Family: GNAT |
N-acetyltransferase, putative [Ricinus communis] |
| 6 |
Hb_010174_130 |
0.0989828382 |
- |
- |
PREDICTED: regulator of G-protein signaling 1 [Jatropha curcas] |
| 7 |
Hb_003404_030 |
0.099312122 |
- |
- |
PREDICTED: monogalactosyldiacylglycerol synthase 2, chloroplastic-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_178968_150 |
0.1018999577 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 9 |
Hb_001476_100 |
0.1071888154 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_007766_010 |
0.1073819025 |
- |
- |
hypothetical protein JCGZ_06718 [Jatropha curcas] |
| 11 |
Hb_004724_040 |
0.1111864161 |
- |
- |
PREDICTED: uncharacterized protein LOC105640153 isoform X1 [Jatropha curcas] |
| 12 |
Hb_010174_120 |
0.1117801352 |
- |
- |
PREDICTED: very-long-chain 3-oxoacyl-CoA reductase 1-like [Gossypium raimondii] |
| 13 |
Hb_002067_160 |
0.1145948888 |
- |
- |
Protein MLO, putative [Ricinus communis] |
| 14 |
Hb_001047_020 |
0.1149009895 |
- |
- |
- |
| 15 |
Hb_002506_010 |
0.1170757115 |
- |
- |
choline transporter-related family protein [Populus trichocarpa] |
| 16 |
Hb_000084_310 |
0.1192580463 |
- |
- |
PREDICTED: rho GTPase-activating protein 4-like [Jatropha curcas] |
| 17 |
Hb_003428_110 |
0.1199188385 |
- |
- |
ARF2-B: ADP-ribosylation factor 2-B [Gossypium arboreum] |
| 18 |
Hb_004738_090 |
0.1202212066 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000784_090 |
0.1208826328 |
- |
- |
PREDICTED: receptor homology region, transmembrane domain- and RING domain-containing protein 2 [Jatropha curcas] |
| 20 |
Hb_001673_120 |
0.1222533033 |
- |
- |
PREDICTED: uncharacterized protein LOC105635794 [Jatropha curcas] |