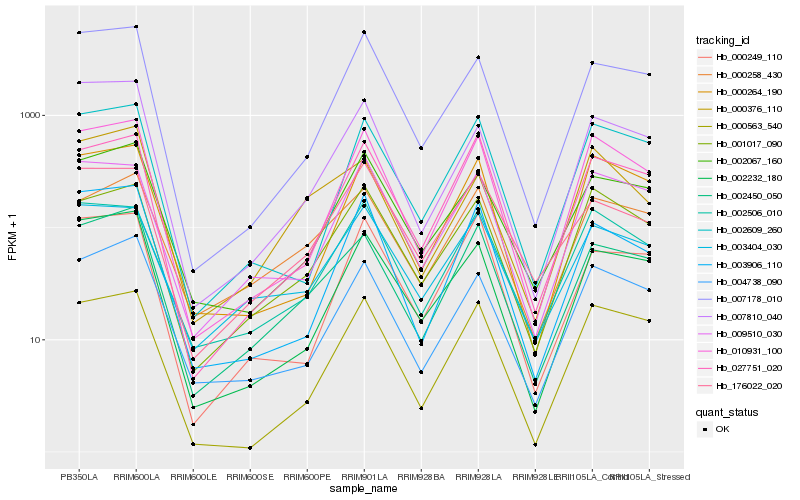
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002450_050 |
0.0 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like [Solanum lycopersicum] |
| 2 |
Hb_000258_430 |
0.0873634119 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 3 |
Hb_004738_090 |
0.0978597727 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_027751_020 |
0.1034393153 |
- |
- |
PREDICTED: uncharacterized protein LOC104886652 [Beta vulgaris subsp. vulgaris] |
| 5 |
Hb_002067_160 |
0.1059179701 |
- |
- |
Protein MLO, putative [Ricinus communis] |
| 6 |
Hb_002232_180 |
0.1073441057 |
- |
- |
PREDICTED: DUF21 domain-containing protein At2g14520-like isoform X1 [Populus euphratica] |
| 7 |
Hb_003404_030 |
0.1106792975 |
- |
- |
PREDICTED: monogalactosyldiacylglycerol synthase 2, chloroplastic-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_010931_100 |
0.112757858 |
- |
- |
PREDICTED: uncharacterized protein LOC105638669 [Jatropha curcas] |
| 9 |
Hb_001017_090 |
0.115065759 |
- |
- |
- |
| 10 |
Hb_002609_260 |
0.1176564217 |
- |
- |
CCCH-type zinc finger protein [Hevea brasiliensis] |
| 11 |
Hb_002506_010 |
0.1196560638 |
- |
- |
choline transporter-related family protein [Populus trichocarpa] |
| 12 |
Hb_000264_190 |
0.1197531048 |
- |
- |
sphingolipid delta 4 desaturase/C-4 hydroxylase protein des2, putative [Ricinus communis] |
| 13 |
Hb_007178_010 |
0.120926708 |
- |
- |
pseudo-hevein [Hevea brasiliensis] |
| 14 |
Hb_003906_110 |
0.1226629772 |
- |
- |
PREDICTED: molybdenum cofactor sulfurase [Jatropha curcas] |
| 15 |
Hb_000249_110 |
0.1231981811 |
- |
- |
hypothetical protein JCGZ_22462 [Jatropha curcas] |
| 16 |
Hb_176022_020 |
0.1232191474 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 17 |
Hb_007810_040 |
0.1251371066 |
- |
- |
PREDICTED: dolichyl-phosphate beta-glucosyltransferase [Jatropha curcas] |
| 18 |
Hb_000563_540 |
0.1272020051 |
- |
- |
PREDICTED: Fanconi anemia group I protein [Jatropha curcas] |
| 19 |
Hb_000376_110 |
0.1284514652 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like [Brachypodium distachyon] |
| 20 |
Hb_009510_030 |
0.1290092762 |
- |
- |
PREDICTED: phosphoinositide phospholipase C 2-like [Jatropha curcas] |