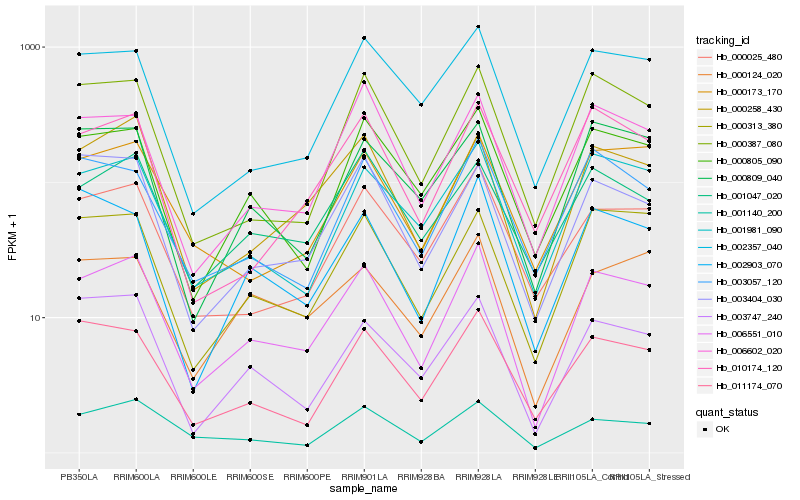
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006551_010 |
0.0 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 2 |
Hb_000258_430 |
0.0928034816 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 3 |
Hb_000313_380 |
0.1035865746 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001981_090 |
0.1069318794 |
- |
- |
PREDICTED: nucleolar GTP-binding protein 1-like [Prunus mume] |
| 5 |
Hb_000805_090 |
0.1107990523 |
- |
- |
hypothetical protein POPTR_0009s03890g [Populus trichocarpa] |
| 6 |
Hb_001140_200 |
0.1114648726 |
- |
- |
PREDICTED: uncharacterized protein LOC105648845 [Jatropha curcas] |
| 7 |
Hb_011174_070 |
0.111781254 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g06540-like [Jatropha curcas] |
| 8 |
Hb_000387_080 |
0.1132256425 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 isoform X2 [Jatropha curcas] |
| 9 |
Hb_003747_240 |
0.1135255521 |
transcription factor |
TF Family: RB |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_006602_020 |
0.1135498119 |
- |
- |
unknown [Medicago truncatula] |
| 11 |
Hb_000025_480 |
0.1141501389 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein 10 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000173_170 |
0.1185311779 |
- |
- |
PREDICTED: UPF0613 protein PB24D3.06c [Jatropha curcas] |
| 13 |
Hb_003404_030 |
0.1187696638 |
- |
- |
PREDICTED: monogalactosyldiacylglycerol synthase 2, chloroplastic-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_002357_040 |
0.1189291349 |
- |
- |
latex-abundant protein [Hevea brasiliensis] |
| 15 |
Hb_000809_040 |
0.1211723111 |
- |
- |
Serine/threonine-protein kinase SAPK10, putative [Ricinus communis] |
| 16 |
Hb_002903_070 |
0.1217825488 |
transcription factor |
TF Family: C2C2-Dof |
zinc finger protein, putative [Ricinus communis] |
| 17 |
Hb_001047_020 |
0.1227127668 |
- |
- |
- |
| 18 |
Hb_000124_020 |
0.1227741849 |
- |
- |
PREDICTED: uncharacterized protein LOC105636589 [Jatropha curcas] |
| 19 |
Hb_010174_120 |
0.1228602168 |
- |
- |
PREDICTED: very-long-chain 3-oxoacyl-CoA reductase 1-like [Gossypium raimondii] |
| 20 |
Hb_003057_120 |
0.1252564429 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |