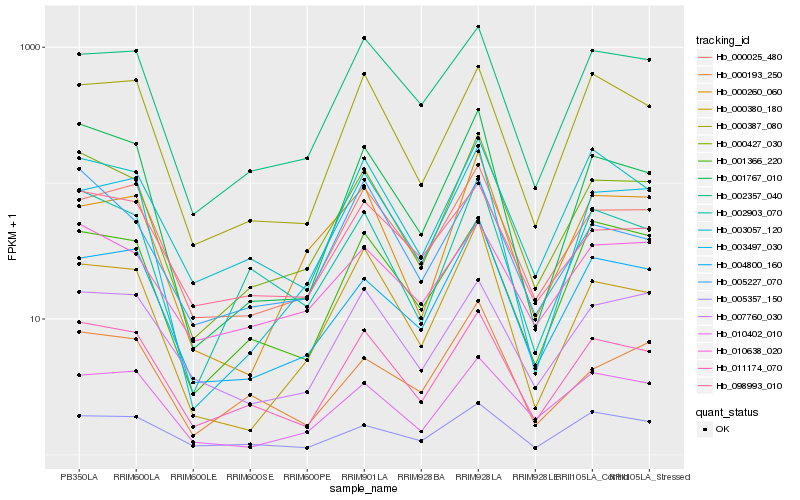
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000427_030 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_011174_070 |
0.0781624296 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g06540-like [Jatropha curcas] |
| 3 |
Hb_002903_070 |
0.0911519922 |
transcription factor |
TF Family: C2C2-Dof |
zinc finger protein, putative [Ricinus communis] |
| 4 |
Hb_001767_010 |
0.0929139738 |
- |
- |
PREDICTED: probable RNA helicase SDE3 [Populus euphratica] |
| 5 |
Hb_004800_160 |
0.1042323585 |
- |
- |
Uncharacterized protein TCM_000403 [Theobroma cacao] |
| 6 |
Hb_000025_480 |
0.1057185445 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein 10 isoform X1 [Jatropha curcas] |
| 7 |
Hb_003057_120 |
0.1080447347 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 8 |
Hb_002357_040 |
0.1103029487 |
- |
- |
latex-abundant protein [Hevea brasiliensis] |
| 9 |
Hb_010638_020 |
0.1128356313 |
- |
- |
PREDICTED: uncharacterized protein LOC105638189 isoform X3 [Jatropha curcas] |
| 10 |
Hb_001366_220 |
0.1155898246 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000380_180 |
0.1171006676 |
- |
- |
kinase, putative [Ricinus communis] |
| 12 |
Hb_007760_030 |
0.1173044542 |
transcription factor |
TF Family: M-type |
hypothetical protein JCGZ_15623 [Jatropha curcas] |
| 13 |
Hb_010402_010 |
0.1178046843 |
- |
- |
BnaC06g23470D [Brassica napus] |
| 14 |
Hb_000387_080 |
0.1193632113 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 isoform X2 [Jatropha curcas] |
| 15 |
Hb_098993_010 |
0.1201030436 |
- |
- |
PREDICTED: chaperone protein dnaJ 72 isoform X2 [Jatropha curcas] |
| 16 |
Hb_003497_030 |
0.1204635595 |
- |
- |
ADP-ribosylation factor, putative [Ricinus communis] |
| 17 |
Hb_000193_250 |
0.1215052089 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g61400 [Jatropha curcas] |
| 18 |
Hb_005227_070 |
0.1229360708 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 19 |
Hb_000260_060 |
0.1256954879 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 6 [Jatropha curcas] |
| 20 |
Hb_005357_150 |
0.1261512236 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |