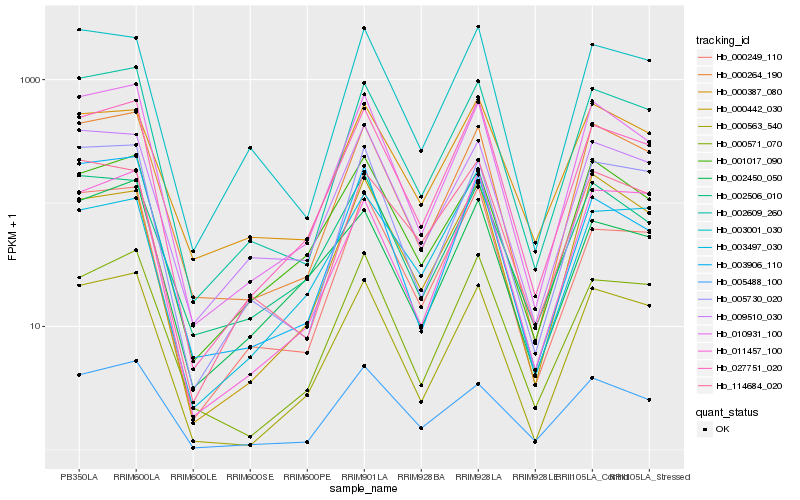
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027751_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104886652 [Beta vulgaris subsp. vulgaris] |
| 2 |
Hb_000249_110 |
0.0713703532 |
- |
- |
hypothetical protein JCGZ_22462 [Jatropha curcas] |
| 3 |
Hb_000571_070 |
0.0740697517 |
transcription factor |
TF Family: G2-like |
PREDICTED: putative Myb family transcription factor At1g14600 [Jatropha curcas] |
| 4 |
Hb_002609_260 |
0.0754757696 |
- |
- |
CCCH-type zinc finger protein [Hevea brasiliensis] |
| 5 |
Hb_010931_100 |
0.0783026126 |
- |
- |
PREDICTED: uncharacterized protein LOC105638669 [Jatropha curcas] |
| 6 |
Hb_000563_540 |
0.0814897833 |
- |
- |
PREDICTED: Fanconi anemia group I protein [Jatropha curcas] |
| 7 |
Hb_000264_190 |
0.0866957607 |
- |
- |
sphingolipid delta 4 desaturase/C-4 hydroxylase protein des2, putative [Ricinus communis] |
| 8 |
Hb_011457_100 |
0.0957520012 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 9 |
Hb_003001_030 |
0.1008908027 |
- |
- |
PREDICTED: putative G3BP-like protein [Jatropha curcas] |
| 10 |
Hb_009510_030 |
0.102051901 |
- |
- |
PREDICTED: phosphoinositide phospholipase C 2-like [Jatropha curcas] |
| 11 |
Hb_005488_100 |
0.1022389089 |
- |
- |
hypothetical protein JCGZ_00295 [Jatropha curcas] |
| 12 |
Hb_001017_090 |
0.1028830692 |
- |
- |
- |
| 13 |
Hb_002450_050 |
0.1034393153 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like [Solanum lycopersicum] |
| 14 |
Hb_003497_030 |
0.1047771464 |
- |
- |
ADP-ribosylation factor, putative [Ricinus communis] |
| 15 |
Hb_000387_080 |
0.1054863066 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 isoform X2 [Jatropha curcas] |
| 16 |
Hb_005730_020 |
0.1060493154 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF3.1-like [Jatropha curcas] |
| 17 |
Hb_002506_010 |
0.1075222764 |
- |
- |
choline transporter-related family protein [Populus trichocarpa] |
| 18 |
Hb_000442_030 |
0.1078165303 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_003906_110 |
0.1105031338 |
- |
- |
PREDICTED: molybdenum cofactor sulfurase [Jatropha curcas] |
| 20 |
Hb_114684_020 |
0.1108667804 |
- |
- |
beta-1,3-glucanase [Hevea brasiliensis] |