| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
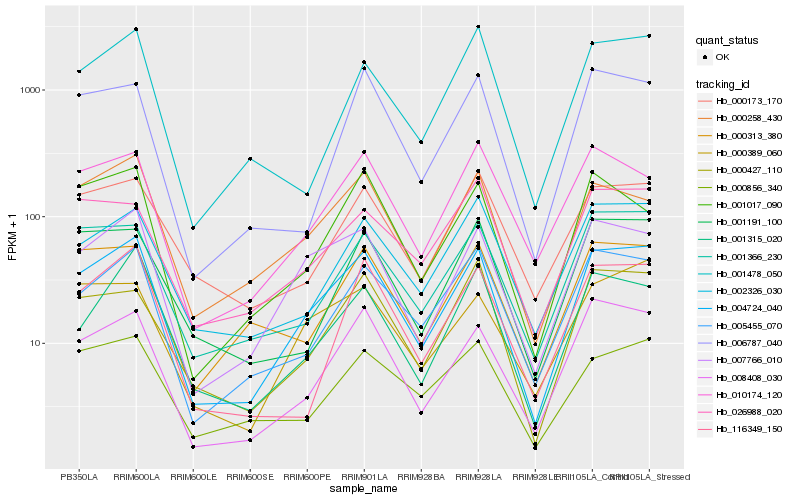
Hb_004724_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640153 isoform X1 [Jatropha curcas] |
| 2 |
Hb_007766_010 |
0.0904557394 |
- |
- |
hypothetical protein JCGZ_06718 [Jatropha curcas] |
| 3 |
Hb_008408_030 |
0.0980435128 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 2 [Jatropha curcas] |
| 4 |
Hb_005455_070 |
0.1054479997 |
- |
- |
hypothetical protein POPTR_0001s04440g [Populus trichocarpa] |
| 5 |
Hb_000258_430 |
0.1111864161 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 6 |
Hb_002326_030 |
0.114506375 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein REM16-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_001366_230 |
0.1171148459 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000427_110 |
0.1171231962 |
- |
- |
PREDICTED: DNA repair protein XRCC2 homolog isoform X2 [Jatropha curcas] |
| 9 |
Hb_010174_120 |
0.121695328 |
- |
- |
PREDICTED: very-long-chain 3-oxoacyl-CoA reductase 1-like [Gossypium raimondii] |
| 10 |
Hb_116349_150 |
0.1223883373 |
- |
- |
PREDICTED: microtubule-associated protein TORTIFOLIA1 [Jatropha curcas] |
| 11 |
Hb_001315_020 |
0.1231471167 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 12 |
Hb_000173_170 |
0.1239406604 |
- |
- |
PREDICTED: UPF0613 protein PB24D3.06c [Jatropha curcas] |
| 13 |
Hb_000856_340 |
0.1244091389 |
- |
- |
PREDICTED: uncharacterized protein LOC105640501 [Jatropha curcas] |
| 14 |
Hb_001191_100 |
0.1262476234 |
- |
- |
F-actin capping protein beta subunit [Populus trichocarpa] |
| 15 |
Hb_000389_060 |
0.127719243 |
- |
- |
PREDICTED: synaptotagmin-3 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000313_380 |
0.1280117056 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001017_090 |
0.1285718364 |
- |
- |
- |
| 18 |
Hb_001478_050 |
0.1303387319 |
- |
- |
PREDICTED: uncharacterized protein LOC105640307 [Jatropha curcas] |
| 19 |
Hb_006787_040 |
0.1307700639 |
- |
- |
polyadenylate-binding protein, putative [Ricinus communis] |
| 20 |
Hb_026988_020 |
0.1311136306 |
- |
- |
outward rectifying K+ channel [Hevea brasiliensis] |