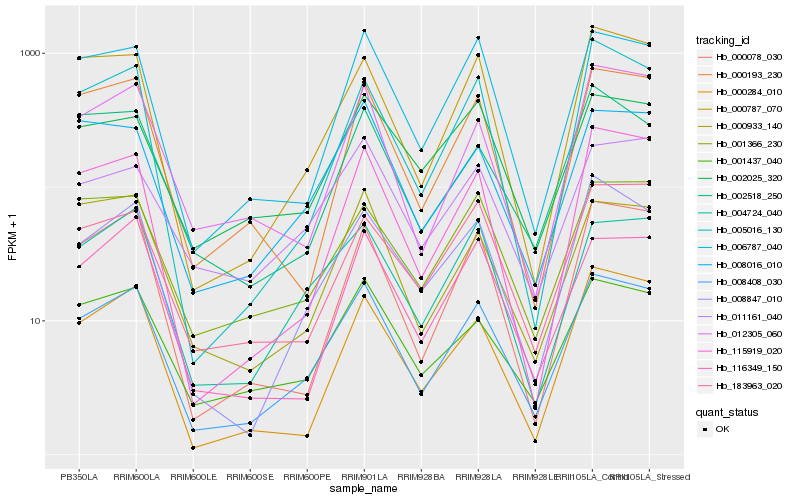
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008408_030 |
0.0 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 2 [Jatropha curcas] |
| 2 |
Hb_008847_010 |
0.0924772936 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 3 |
Hb_006787_040 |
0.0929518483 |
- |
- |
polyadenylate-binding protein, putative [Ricinus communis] |
| 4 |
Hb_001437_040 |
0.093976935 |
- |
- |
PREDICTED: ATPase GET3-like [Vitis vinifera] |
| 5 |
Hb_000787_070 |
0.0965915885 |
- |
- |
Early nodulin 55-2 precursor, putative [Ricinus communis] |
| 6 |
Hb_004724_040 |
0.0980435128 |
- |
- |
PREDICTED: uncharacterized protein LOC105640153 isoform X1 [Jatropha curcas] |
| 7 |
Hb_115919_020 |
0.1019325466 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranyl-diphosphate synthase [Hevea brasiliensis] |
| 8 |
Hb_008016_010 |
0.1023103787 |
- |
- |
PREDICTED: protein LURP-one-related 15-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_000078_030 |
0.1064535812 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_012305_060 |
0.106977069 |
- |
- |
protein translocase, putative [Ricinus communis] |
| 11 |
Hb_000193_230 |
0.108418699 |
- |
- |
PREDICTED: NEP1-interacting protein-like 2 isoform X1 [Jatropha curcas] |
| 12 |
Hb_005016_130 |
0.1106761616 |
- |
- |
PREDICTED: uncharacterized protein LOC105642243 [Jatropha curcas] |
| 13 |
Hb_000933_140 |
0.1112816849 |
- |
- |
PREDICTED: uncharacterized protein LOC105630501 [Jatropha curcas] |
| 14 |
Hb_116349_150 |
0.111558136 |
- |
- |
PREDICTED: microtubule-associated protein TORTIFOLIA1 [Jatropha curcas] |
| 15 |
Hb_001366_230 |
0.1116880249 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_011161_040 |
0.1130140974 |
- |
- |
PREDICTED: uncharacterized protein LOC105638905 isoform X2 [Jatropha curcas] |
| 17 |
Hb_183963_020 |
0.1170758448 |
- |
- |
PREDICTED: uncharacterized protein LOC105644615 [Jatropha curcas] |
| 18 |
Hb_002518_250 |
0.1172652319 |
- |
- |
conserved hypothetical protein 10 [Hevea brasiliensis] |
| 19 |
Hb_000284_010 |
0.1182238376 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_04974 [Jatropha curcas] |
| 20 |
Hb_002025_320 |
0.1185553642 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit I-like [Jatropha curcas] |