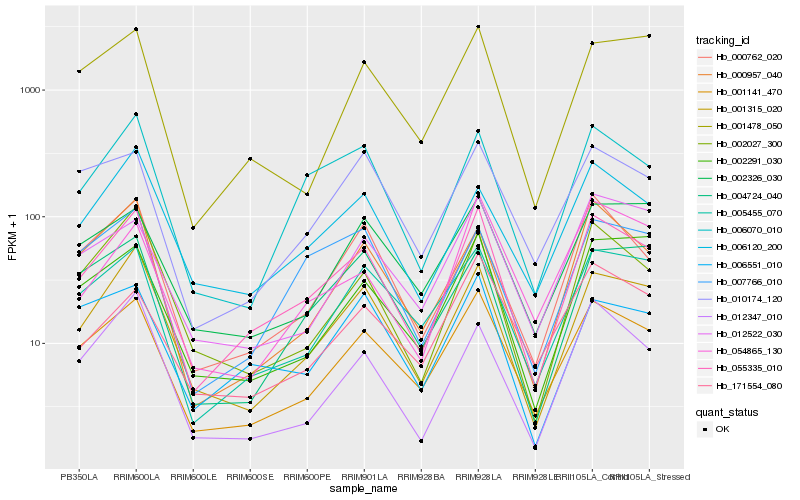
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_055335_010 |
0.0 |
- |
- |
PREDICTED: tetraspanin-2 [Jatropha curcas] |
| 2 |
Hb_001141_470 |
0.0869623384 |
- |
- |
PREDICTED: uncharacterized protein LOC101254423 [Solanum lycopersicum] |
| 3 |
Hb_001315_020 |
0.0978031256 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 4 |
Hb_006070_010 |
0.1080324658 |
rubber biosynthesis |
Gene Name: CPT2 |
cis-prenyltransferase 3 [Hevea brasiliensis] |
| 5 |
Hb_005455_070 |
0.1178518467 |
- |
- |
hypothetical protein POPTR_0001s04440g [Populus trichocarpa] |
| 6 |
Hb_002027_300 |
0.1231481424 |
- |
- |
PREDICTED: rho GTPase-activating protein 2 [Jatropha curcas] |
| 7 |
Hb_000957_040 |
0.1252276482 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RNF144A-B [Jatropha curcas] |
| 8 |
Hb_006120_200 |
0.1319922371 |
- |
- |
PREDICTED: uncharacterized protein LOC103333282 [Prunus mume] |
| 9 |
Hb_007766_010 |
0.1368423651 |
- |
- |
hypothetical protein JCGZ_06718 [Jatropha curcas] |
| 10 |
Hb_006551_010 |
0.1370507222 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 11 |
Hb_000762_020 |
0.1370738559 |
- |
- |
PREDICTED: uncharacterized protein LOC105641515 [Jatropha curcas] |
| 12 |
Hb_010174_120 |
0.1384830359 |
- |
- |
PREDICTED: very-long-chain 3-oxoacyl-CoA reductase 1-like [Gossypium raimondii] |
| 13 |
Hb_012347_010 |
0.1385428555 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_03155 [Jatropha curcas] |
| 14 |
Hb_002291_030 |
0.1405254508 |
- |
- |
PREDICTED: vesicle transport protein SFT2B [Jatropha curcas] |
| 15 |
Hb_001478_050 |
0.1411521203 |
- |
- |
PREDICTED: uncharacterized protein LOC105640307 [Jatropha curcas] |
| 16 |
Hb_012522_030 |
0.1426907152 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_171554_080 |
0.1429412222 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 18 |
Hb_002326_030 |
0.1443847578 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein REM16-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_054865_130 |
0.1462805413 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 20 |
Hb_004724_040 |
0.147205857 |
- |
- |
PREDICTED: uncharacterized protein LOC105640153 isoform X1 [Jatropha curcas] |