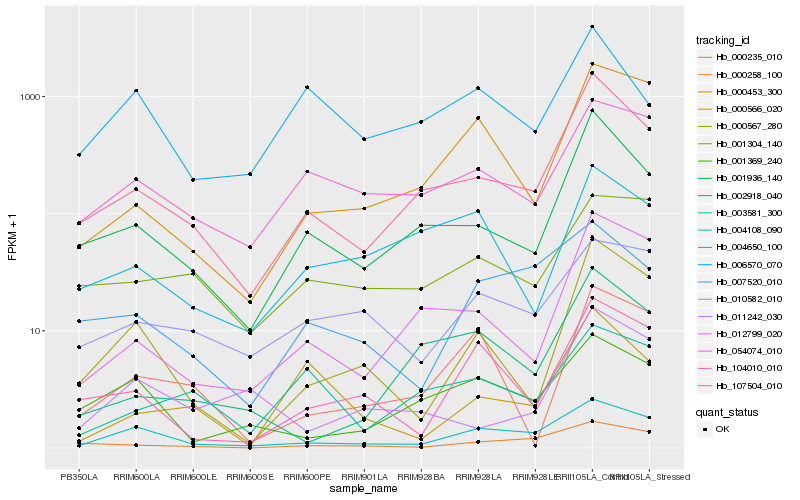
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_107504_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_001936_140 |
0.0684977423 |
- |
- |
PREDICTED: ATP-citrate synthase alpha chain protein 1 [Jatropha curcas] |
| 3 |
Hb_012799_020 |
0.1429769731 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 4 |
Hb_002918_040 |
0.1801339178 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 5 |
Hb_004108_090 |
0.1863027733 |
- |
- |
PREDICTED: uncharacterized protein LOC105650231 [Jatropha curcas] |
| 6 |
Hb_054074_010 |
0.1930512346 |
- |
- |
PREDICTED: uncharacterized protein LOC105645601 [Jatropha curcas] |
| 7 |
Hb_000453_300 |
0.1956995003 |
- |
- |
latex cystatin [Hevea brasiliensis] |
| 8 |
Hb_000567_280 |
0.19890769 |
- |
- |
PREDICTED: uncharacterized protein At4g06598-like [Populus euphratica] |
| 9 |
Hb_000258_100 |
0.2032378662 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_004650_100 |
0.2094515662 |
- |
- |
Ubiquitin-fold modifier 1 precursor, putative [Ricinus communis] |
| 11 |
Hb_001369_240 |
0.215919366 |
- |
- |
- |
| 12 |
Hb_001304_140 |
0.2200249514 |
- |
- |
PREDICTED: transmembrane protein 256 homolog [Jatropha curcas] |
| 13 |
Hb_003581_300 |
0.2212784351 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105630404 isoform X1 [Jatropha curcas] |
| 14 |
Hb_007520_010 |
0.2223562705 |
- |
- |
PREDICTED: uncharacterized protein LOC105640383 [Jatropha curcas] |
| 15 |
Hb_104010_010 |
0.2225945534 |
- |
- |
hypothetical protein JCGZ_26423 [Jatropha curcas] |
| 16 |
Hb_011242_030 |
0.231157129 |
- |
- |
- |
| 17 |
Hb_000235_010 |
0.2339043094 |
- |
- |
PREDICTED: serine carboxypeptidase-like 18 [Jatropha curcas] |
| 18 |
Hb_010582_010 |
0.2339377146 |
- |
- |
hypothetical protein POPTR_0011s15670g [Populus trichocarpa] |
| 19 |
Hb_006570_070 |
0.2354973774 |
- |
- |
cyclophilin [Hevea brasiliensis] |
| 20 |
Hb_000566_020 |
0.2355812416 |
- |
- |
hypothetical protein POPTR_0001s13520g [Populus trichocarpa] |