| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
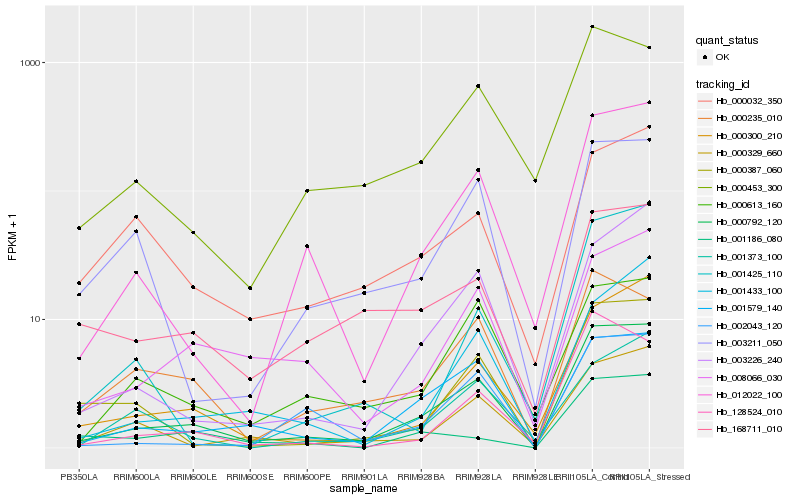
Hb_000792_120 |
0.0 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 1 isoform X2 [Jatropha curcas] |
| 2 |
Hb_012022_100 |
0.1374428547 |
- |
- |
CPRD49 family protein [Populus trichocarpa] |
| 3 |
Hb_001579_140 |
0.1395927911 |
- |
- |
PREDICTED: probable inactive poly [ADP-ribose] polymerase SRO2 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000300_210 |
0.1516693308 |
- |
- |
gd2b, putative [Ricinus communis] |
| 5 |
Hb_000329_660 |
0.1606299333 |
- |
- |
PREDICTED: carotenoid cleavage dioxygenase 7, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000387_060 |
0.164560541 |
- |
- |
- |
| 7 |
Hb_008066_030 |
0.1700888797 |
- |
- |
PREDICTED: uncharacterized protein LOC105631333 [Jatropha curcas] |
| 8 |
Hb_003226_240 |
0.1702264422 |
- |
- |
PREDICTED: caffeoylshikimate esterase [Jatropha curcas] |
| 9 |
Hb_002043_120 |
0.1707194566 |
- |
- |
protein with unknown function [Ricinus communis] |
| 10 |
Hb_000453_300 |
0.1713111736 |
- |
- |
latex cystatin [Hevea brasiliensis] |
| 11 |
Hb_128524_010 |
0.17613655 |
- |
- |
PREDICTED: polyadenylate-binding protein 7 isoform X1 [Jatropha curcas] |
| 12 |
Hb_003211_050 |
0.176619922 |
- |
- |
PREDICTED: RNA-binding protein 24-B isoform X1 [Jatropha curcas] |
| 13 |
Hb_000235_010 |
0.1779435283 |
- |
- |
PREDICTED: serine carboxypeptidase-like 18 [Jatropha curcas] |
| 14 |
Hb_000613_160 |
0.1792919455 |
- |
- |
coated vesicle membrane protein, putative [Ricinus communis] |
| 15 |
Hb_001433_100 |
0.1821349374 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 3 isoform X1 [Jatropha curcas] |
| 16 |
Hb_168711_010 |
0.182242152 |
- |
- |
PREDICTED: (DL)-glycerol-3-phosphatase 2 [Sesamum indicum] |
| 17 |
Hb_000032_350 |
0.1878043172 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001373_100 |
0.1901895447 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001186_080 |
0.198390502 |
- |
- |
PREDICTED: F-box protein FBW2-like [Nelumbo nucifera] |
| 20 |
Hb_001425_110 |
0.1987737204 |
- |
- |
hypothetical protein Csa_6G106200 [Cucumis sativus] |