| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
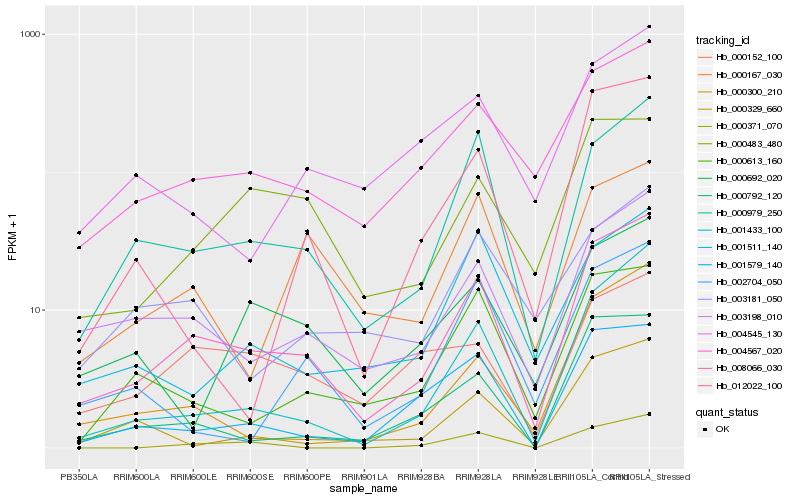
Hb_008066_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631333 [Jatropha curcas] |
| 2 |
Hb_000979_250 |
0.1272737944 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_003198_010 |
0.1332074097 |
- |
- |
Valine--tRNA ligase [Gossypium arboreum] |
| 4 |
Hb_004567_020 |
0.1512048252 |
- |
- |
Small ubiquitin-related modifier 2 [Morus notabilis] |
| 5 |
Hb_001433_100 |
0.1520426438 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 3 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000792_120 |
0.1700888797 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 1 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000300_210 |
0.178214608 |
- |
- |
gd2b, putative [Ricinus communis] |
| 8 |
Hb_000613_160 |
0.1797043476 |
- |
- |
coated vesicle membrane protein, putative [Ricinus communis] |
| 9 |
Hb_001579_140 |
0.1802821542 |
- |
- |
PREDICTED: probable inactive poly [ADP-ribose] polymerase SRO2 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000329_660 |
0.1886400661 |
- |
- |
PREDICTED: carotenoid cleavage dioxygenase 7, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000483_480 |
0.1895057904 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_004545_130 |
0.1916032766 |
- |
- |
uncharacterized protein LOC100305465 [Glycine max] |
| 13 |
Hb_000167_030 |
0.1916973358 |
- |
- |
PREDICTED: uncharacterized protein LOC105641111 isoform X1 [Jatropha curcas] |
| 14 |
Hb_003181_050 |
0.1919513554 |
- |
- |
PREDICTED: DNA replication complex GINS protein PSF2 [Jatropha curcas] |
| 15 |
Hb_001511_140 |
0.1939602646 |
- |
- |
PREDICTED: RING-H2 finger protein ATL66 [Jatropha curcas] |
| 16 |
Hb_000371_070 |
0.1952819066 |
- |
- |
PREDICTED: uncharacterized protein LOC103863319 [Brassica rapa] |
| 17 |
Hb_000152_100 |
0.196530251 |
- |
- |
PREDICTED: probable protein phosphatase 2C 75 isoform X2 [Jatropha curcas] |
| 18 |
Hb_012022_100 |
0.1975148234 |
- |
- |
CPRD49 family protein [Populus trichocarpa] |
| 19 |
Hb_000692_020 |
0.19817158 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase ATL23 [Jatropha curcas] |
| 20 |
Hb_002704_050 |
0.198946626 |
- |
- |
cysteine-type peptidase, putative [Ricinus communis] |