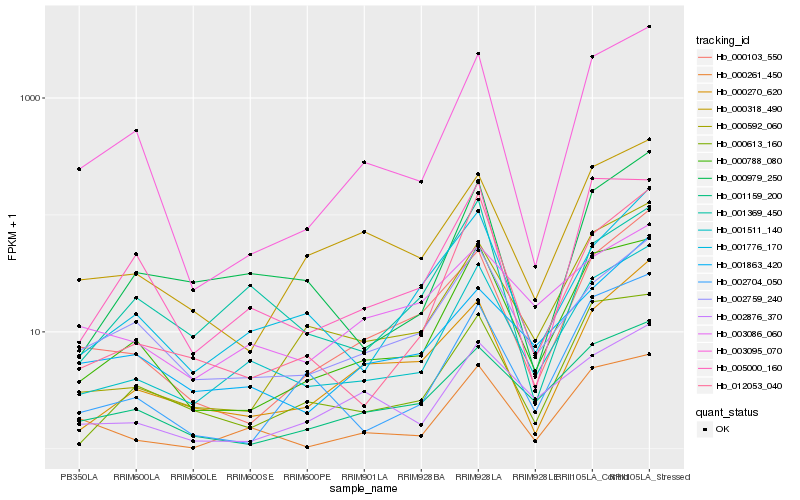
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001511_140 |
0.0 |
- |
- |
PREDICTED: RING-H2 finger protein ATL66 [Jatropha curcas] |
| 2 |
Hb_000788_080 |
0.1196786044 |
- |
- |
PREDICTED: uncharacterized protein LOC105629703 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001776_170 |
0.1283608536 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000979_250 |
0.1311650895 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_012053_040 |
0.1446363228 |
- |
- |
PREDICTED: putative phospholipid:diacylglycerol acyltransferase 2 [Jatropha curcas] |
| 6 |
Hb_002759_240 |
0.1464319223 |
- |
- |
hypothetical protein EUGRSUZ_E01319 [Eucalyptus grandis] |
| 7 |
Hb_000592_060 |
0.1478118628 |
- |
- |
PREDICTED: ras-related protein RABA6a [Populus euphratica] |
| 8 |
Hb_000261_450 |
0.1481161709 |
- |
- |
Pectinesterase U1 precursor, putative [Ricinus communis] |
| 9 |
Hb_001159_200 |
0.1532771073 |
- |
- |
PREDICTED: clp protease-related protein At4g12060, chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_000613_160 |
0.1543814111 |
- |
- |
coated vesicle membrane protein, putative [Ricinus communis] |
| 11 |
Hb_000103_550 |
0.1582109228 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit gamma 2 isoform X1 [Jatropha curcas] |
| 12 |
Hb_003095_070 |
0.1604392652 |
- |
- |
hypothetical protein POPTR_0009s11230g [Populus trichocarpa] |
| 13 |
Hb_000318_490 |
0.1606605677 |
- |
- |
PREDICTED: mitochondrial import receptor subunit TOM6 homolog [Jatropha curcas] |
| 14 |
Hb_001863_420 |
0.161168175 |
- |
- |
PREDICTED: probable protein phosphatase 2C 10, partial [Vitis vinifera] |
| 15 |
Hb_000270_620 |
0.1619258331 |
- |
- |
PREDICTED: succinate dehydrogenase subunit 7B, mitochondrial-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_002876_370 |
0.1631968337 |
- |
- |
hypothetical protein B456_013G140400 [Gossypium raimondii] |
| 17 |
Hb_002704_050 |
0.1659129093 |
- |
- |
cysteine-type peptidase, putative [Ricinus communis] |
| 18 |
Hb_003086_060 |
0.1662557102 |
- |
- |
PREDICTED: uncharacterized protein LOC105629937 [Jatropha curcas] |
| 19 |
Hb_001369_450 |
0.1667401276 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 20 |
Hb_005000_160 |
0.1670963207 |
- |
- |
PREDICTED: uncharacterized protein LOC105637882 [Jatropha curcas] |