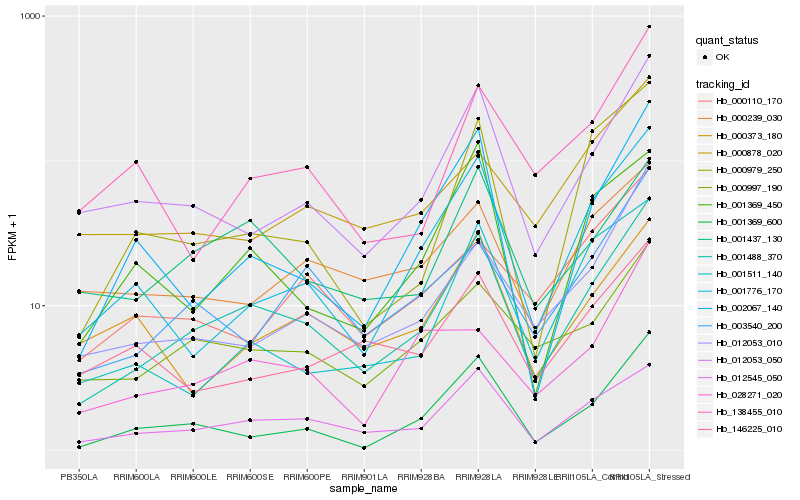
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001488_370 |
0.0 |
- |
- |
Esterase PIR7B, putative [Ricinus communis] |
| 2 |
Hb_012545_050 |
0.114583572 |
- |
- |
PREDICTED: serine carboxypeptidase II-3-like [Jatropha curcas] |
| 3 |
Hb_012053_050 |
0.1357168549 |
- |
- |
PREDICTED: UDP-D-apiose/UDP-D-xylose synthase 2 [Jatropha curcas] |
| 4 |
Hb_001369_600 |
0.1439966988 |
- |
- |
kinase, putative [Ricinus communis] |
| 5 |
Hb_000997_190 |
0.1459494729 |
- |
- |
PREDICTED: magnesium transporter MRS2-I-like [Jatropha curcas] |
| 6 |
Hb_001437_130 |
0.1467155205 |
- |
- |
hypothetical protein F383_15788 [Gossypium arboreum] |
| 7 |
Hb_001776_170 |
0.1591434602 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002067_140 |
0.1600425112 |
- |
- |
- |
| 9 |
Hb_003540_200 |
0.163564375 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA3-like [Jatropha curcas] |
| 10 |
Hb_012053_010 |
0.1636191061 |
- |
- |
srpk, putative [Ricinus communis] |
| 11 |
Hb_000979_250 |
0.1646401843 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000239_030 |
0.176267558 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin 2-like [Jatropha curcas] |
| 13 |
Hb_001369_450 |
0.17636494 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 14 |
Hb_138455_010 |
0.1785657757 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 15 |
Hb_000373_180 |
0.1832847627 |
transcription factor |
TF Family: Trihelix |
hypothetical protein JCGZ_25182 [Jatropha curcas] |
| 16 |
Hb_001511_140 |
0.1838318757 |
- |
- |
PREDICTED: RING-H2 finger protein ATL66 [Jatropha curcas] |
| 17 |
Hb_000110_170 |
0.1842901132 |
- |
- |
PREDICTED: arginine biosynthesis bifunctional protein ArgJ, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_146225_010 |
0.1843316608 |
- |
- |
mitochondrial inner membrane protease subunit, putative [Ricinus communis] |
| 19 |
Hb_000878_020 |
0.1843699331 |
- |
- |
PREDICTED: uncharacterized protein LOC105629937 [Jatropha curcas] |
| 20 |
Hb_028271_020 |
0.1861098327 |
- |
- |
acyl-CoA oxidase, putative [Ricinus communis] |