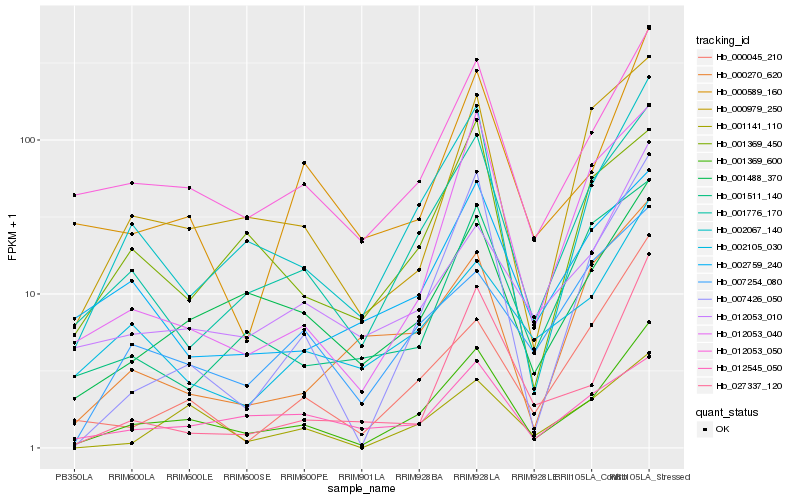
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001369_600 |
0.0 |
- |
- |
kinase, putative [Ricinus communis] |
| 2 |
Hb_002067_140 |
0.1119245537 |
- |
- |
- |
| 3 |
Hb_012053_050 |
0.1206114848 |
- |
- |
PREDICTED: UDP-D-apiose/UDP-D-xylose synthase 2 [Jatropha curcas] |
| 4 |
Hb_001776_170 |
0.1235186399 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001488_370 |
0.1439966988 |
- |
- |
Esterase PIR7B, putative [Ricinus communis] |
| 6 |
Hb_007426_050 |
0.163598033 |
transcription factor |
TF Family: Tify |
JAZ8 [Hevea brasiliensis] |
| 7 |
Hb_000589_160 |
0.1671532574 |
- |
- |
PREDICTED: expansin-A4 [Jatropha curcas] |
| 8 |
Hb_000979_250 |
0.167814054 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001141_110 |
0.1756979447 |
- |
- |
hypothetical protein JCGZ_02206 [Jatropha curcas] |
| 10 |
Hb_012053_010 |
0.1828222739 |
- |
- |
srpk, putative [Ricinus communis] |
| 11 |
Hb_012053_040 |
0.183290122 |
- |
- |
PREDICTED: putative phospholipid:diacylglycerol acyltransferase 2 [Jatropha curcas] |
| 12 |
Hb_002105_030 |
0.1893276528 |
- |
- |
- |
| 13 |
Hb_001369_450 |
0.1900904773 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 14 |
Hb_007254_080 |
0.1902774174 |
- |
- |
- |
| 15 |
Hb_002759_240 |
0.1956414713 |
- |
- |
hypothetical protein EUGRSUZ_E01319 [Eucalyptus grandis] |
| 16 |
Hb_027337_120 |
0.1979218723 |
- |
- |
PREDICTED: uncharacterized protein LOC104430249 [Eucalyptus grandis] |
| 17 |
Hb_000045_210 |
0.1979968921 |
- |
- |
- |
| 18 |
Hb_012545_050 |
0.1985855088 |
- |
- |
PREDICTED: serine carboxypeptidase II-3-like [Jatropha curcas] |
| 19 |
Hb_001511_140 |
0.2029246354 |
- |
- |
PREDICTED: RING-H2 finger protein ATL66 [Jatropha curcas] |
| 20 |
Hb_000270_620 |
0.2032530447 |
- |
- |
PREDICTED: succinate dehydrogenase subunit 7B, mitochondrial-like isoform X1 [Jatropha curcas] |