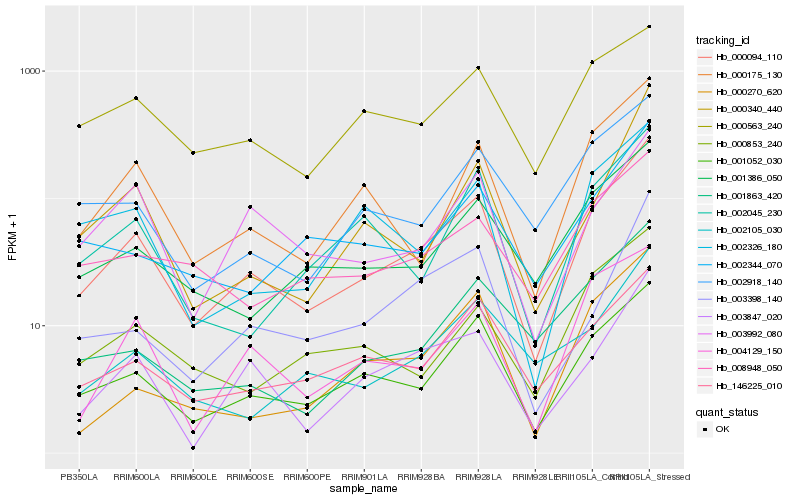
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000094_110 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001052_030 |
0.10204753 |
- |
- |
PREDICTED: protein TIC 20-IV, chloroplastic [Jatropha curcas] |
| 3 |
Hb_003847_020 |
0.1322025453 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 4 |
Hb_000175_130 |
0.1345577841 |
- |
- |
PREDICTED: PTI1-like tyrosine-protein kinase At3g15890 [Jatropha curcas] |
| 5 |
Hb_146225_010 |
0.1392851479 |
- |
- |
mitochondrial inner membrane protease subunit, putative [Ricinus communis] |
| 6 |
Hb_001386_050 |
0.1421827877 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit gamma-like [Jatropha curcas] |
| 7 |
Hb_002918_140 |
0.1497586561 |
- |
- |
PREDICTED: protein yippee-like At5g53940 [Jatropha curcas] |
| 8 |
Hb_004129_150 |
0.1513780181 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-4a-like [Jatropha curcas] |
| 9 |
Hb_002326_180 |
0.1541909636 |
- |
- |
Protein Z, putative [Ricinus communis] |
| 10 |
Hb_002105_030 |
0.1546131584 |
- |
- |
- |
| 11 |
Hb_003398_140 |
0.1548110768 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 12 |
Hb_000340_440 |
0.1554104263 |
- |
- |
PREDICTED: gibberellin receptor GID1B [Jatropha curcas] |
| 13 |
Hb_002045_230 |
0.1556828178 |
- |
- |
Ammonium transporter 2 [Theobroma cacao] |
| 14 |
Hb_008948_050 |
0.1574617309 |
- |
- |
hypothetical protein JCGZ_13339 [Jatropha curcas] |
| 15 |
Hb_002344_070 |
0.1576166824 |
- |
- |
PREDICTED: proteasome subunit alpha type-6 [Jatropha curcas] |
| 16 |
Hb_003992_080 |
0.1588099246 |
- |
- |
PREDICTED: protein LURP-one-related 5 [Jatropha curcas] |
| 17 |
Hb_000563_240 |
0.1596541724 |
- |
- |
PREDICTED: 60S ribosomal protein L19-3 [Jatropha curcas] |
| 18 |
Hb_001863_420 |
0.1604355069 |
- |
- |
PREDICTED: probable protein phosphatase 2C 10, partial [Vitis vinifera] |
| 19 |
Hb_000853_240 |
0.1604940386 |
- |
- |
- |
| 20 |
Hb_000270_620 |
0.165524424 |
- |
- |
PREDICTED: succinate dehydrogenase subunit 7B, mitochondrial-like isoform X1 [Jatropha curcas] |