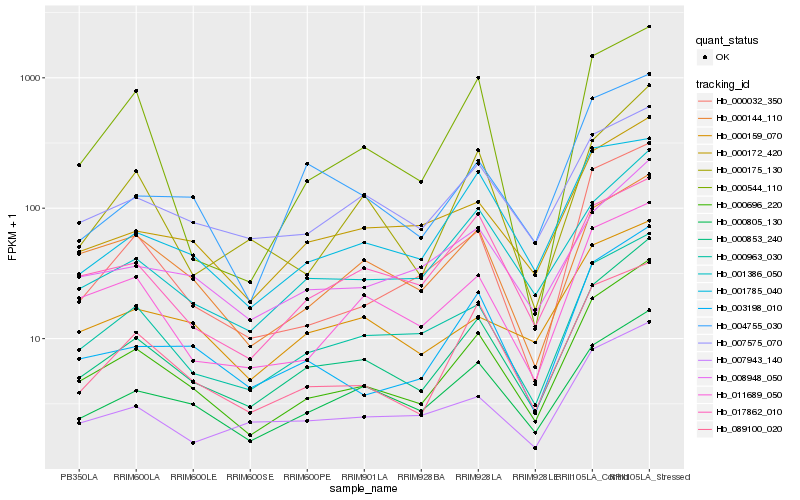
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000696_220 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647113 [Jatropha curcas] |
| 2 |
Hb_000853_240 |
0.0565525035 |
- |
- |
- |
| 3 |
Hb_003198_010 |
0.0981640022 |
- |
- |
Valine--tRNA ligase [Gossypium arboreum] |
| 4 |
Hb_089100_020 |
0.1039122036 |
- |
- |
PREDICTED: cyclin-A1-1 [Jatropha curcas] |
| 5 |
Hb_000032_350 |
0.1057163803 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000963_030 |
0.107706672 |
- |
- |
PREDICTED: uncharacterized protein LOC105628095 [Jatropha curcas] |
| 7 |
Hb_001386_050 |
0.1082609244 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit gamma-like [Jatropha curcas] |
| 8 |
Hb_011689_050 |
0.111213667 |
- |
- |
hypothetical protein POPTR_0008s20290g [Populus trichocarpa] |
| 9 |
Hb_000172_420 |
0.1143340505 |
- |
- |
60S ribosomal protein L30, putative [Ricinus communis] |
| 10 |
Hb_000805_130 |
0.1178547511 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_008948_050 |
0.1225483553 |
- |
- |
hypothetical protein JCGZ_13339 [Jatropha curcas] |
| 12 |
Hb_000175_130 |
0.1311437305 |
- |
- |
PREDICTED: PTI1-like tyrosine-protein kinase At3g15890 [Jatropha curcas] |
| 13 |
Hb_007943_140 |
0.1324969547 |
- |
- |
PREDICTED: uncharacterized protein LOC103340977 [Prunus mume] |
| 14 |
Hb_007575_070 |
0.1325507238 |
- |
- |
Histone H3.2 [Glycine soja] |
| 15 |
Hb_000159_070 |
0.1355429722 |
- |
- |
JHL03K20.4 [Jatropha curcas] |
| 16 |
Hb_017862_010 |
0.1384336705 |
- |
- |
PREDICTED: signal recognition particle 19 kDa protein-like [Jatropha curcas] |
| 17 |
Hb_000144_110 |
0.1396753809 |
- |
- |
hypothetical protein CICLE_v10013189mg [Citrus clementina] |
| 18 |
Hb_001785_040 |
0.1414426856 |
- |
- |
cytochrome b5 isoform Cb5-A [Vernicia fordii] |
| 19 |
Hb_004755_030 |
0.1417040992 |
- |
- |
EG964 [Manihot glaziovii] |
| 20 |
Hb_000544_110 |
0.14214138 |
- |
- |
PREDICTED: probable calcium-binding protein CML27 [Jatropha curcas] |