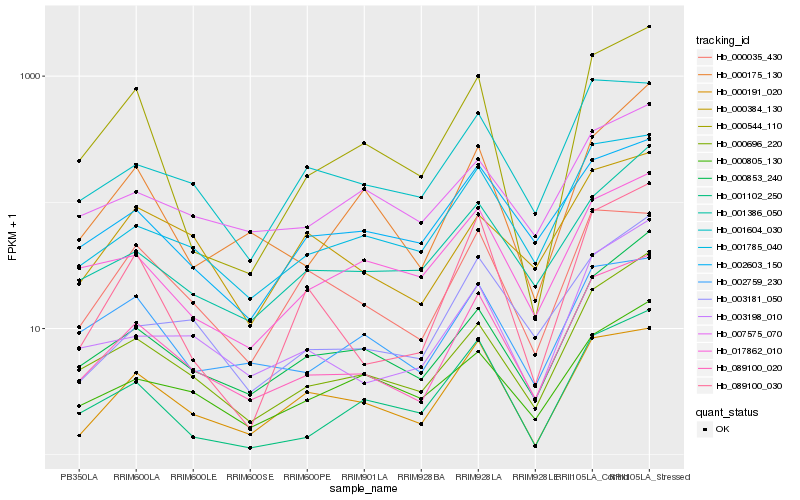
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_089100_020 |
0.0 |
- |
- |
PREDICTED: cyclin-A1-1 [Jatropha curcas] |
| 2 |
Hb_001785_040 |
0.1019641226 |
- |
- |
cytochrome b5 isoform Cb5-A [Vernicia fordii] |
| 3 |
Hb_000696_220 |
0.1039122036 |
- |
- |
PREDICTED: uncharacterized protein LOC105647113 [Jatropha curcas] |
| 4 |
Hb_000853_240 |
0.1212793939 |
- |
- |
- |
| 5 |
Hb_003198_010 |
0.1236273796 |
- |
- |
Valine--tRNA ligase [Gossypium arboreum] |
| 6 |
Hb_000191_020 |
0.1241118602 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003181_050 |
0.1241589688 |
- |
- |
PREDICTED: DNA replication complex GINS protein PSF2 [Jatropha curcas] |
| 8 |
Hb_001604_030 |
0.127769356 |
- |
- |
ubiquitin-conjugating enzyme E2 35-like [Cucumis sativus] |
| 9 |
Hb_001386_050 |
0.1316152698 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit gamma-like [Jatropha curcas] |
| 10 |
Hb_002759_230 |
0.1329970277 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 25 [Jatropha curcas] |
| 11 |
Hb_002603_150 |
0.1333095325 |
- |
- |
PREDICTED: protein yippee-like [Jatropha curcas] |
| 12 |
Hb_017862_010 |
0.1342103416 |
- |
- |
PREDICTED: signal recognition particle 19 kDa protein-like [Jatropha curcas] |
| 13 |
Hb_000805_130 |
0.1357966777 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_007575_070 |
0.137907102 |
- |
- |
Histone H3.2 [Glycine soja] |
| 15 |
Hb_089100_030 |
0.1387563585 |
- |
- |
- |
| 16 |
Hb_000544_110 |
0.1396527456 |
- |
- |
PREDICTED: probable calcium-binding protein CML27 [Jatropha curcas] |
| 17 |
Hb_001102_250 |
0.1398025935 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase CCNB1IP1 homolog isoform X1 [Jatropha curcas] |
| 18 |
Hb_000175_130 |
0.1398508067 |
- |
- |
PREDICTED: PTI1-like tyrosine-protein kinase At3g15890 [Jatropha curcas] |
| 19 |
Hb_000035_430 |
0.1421231315 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2-like [Jatropha curcas] |
| 20 |
Hb_000384_130 |
0.142848392 |
- |
- |
uridylate kinase plant, putative [Ricinus communis] |