| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
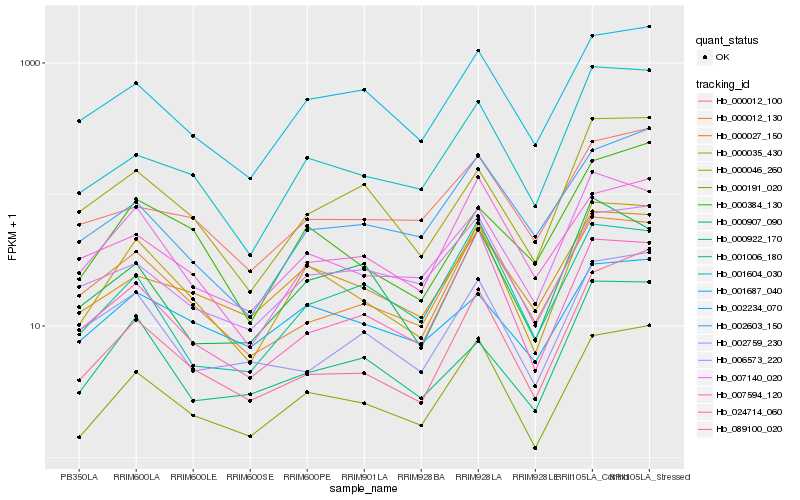
Hb_000035_430 |
0.0 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2-like [Jatropha curcas] |
| 2 |
Hb_000191_020 |
0.0947102317 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_007140_020 |
0.1001414152 |
- |
- |
hypothetical protein CICLE_v10008419mg [Citrus clementina] |
| 4 |
Hb_001687_040 |
0.1195429305 |
- |
- |
actin-depolymerizing factor [Hevea brasiliensis] |
| 5 |
Hb_001604_030 |
0.1206151593 |
- |
- |
ubiquitin-conjugating enzyme E2 35-like [Cucumis sativus] |
| 6 |
Hb_000027_150 |
0.1278512298 |
- |
- |
mitochondrial carrier protein, putative [Ricinus communis] |
| 7 |
Hb_000012_130 |
0.1284094387 |
- |
- |
PREDICTED: uncharacterized protein LOC100807540 isoform X2 [Glycine max] |
| 8 |
Hb_000384_130 |
0.1312071054 |
- |
- |
uridylate kinase plant, putative [Ricinus communis] |
| 9 |
Hb_000046_260 |
0.13315135 |
- |
- |
acyl carrier protein, putative [Ricinus communis] |
| 10 |
Hb_024714_060 |
0.1336127096 |
- |
- |
PREDICTED: thiamine pyrophosphokinase 1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000907_090 |
0.1406605416 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 12 |
Hb_089100_020 |
0.1421231315 |
- |
- |
PREDICTED: cyclin-A1-1 [Jatropha curcas] |
| 13 |
Hb_006573_220 |
0.1457481365 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 1 [Jatropha curcas] |
| 14 |
Hb_000922_170 |
0.1457874106 |
- |
- |
PREDICTED: uncharacterized protein LOC105640383 [Jatropha curcas] |
| 15 |
Hb_001006_180 |
0.1493705407 |
- |
- |
PREDICTED: endonuclease V-like [Jatropha curcas] |
| 16 |
Hb_002603_150 |
0.1494167762 |
- |
- |
PREDICTED: protein yippee-like [Jatropha curcas] |
| 17 |
Hb_002234_070 |
0.1511770197 |
- |
- |
PREDICTED: polynucleotide 5'-hydroxyl-kinase nol9 isoform X1 [Jatropha curcas] |
| 18 |
Hb_007594_120 |
0.1513001799 |
transcription factor |
TF Family: MIKC |
MADS-box transcription factor 2 [Hevea brasiliensis] |
| 19 |
Hb_002759_230 |
0.1518520213 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 25 [Jatropha curcas] |
| 20 |
Hb_000012_100 |
0.1530589798 |
- |
- |
UPF0587 protein C1orf123 homolog [Jatropha curcas] |