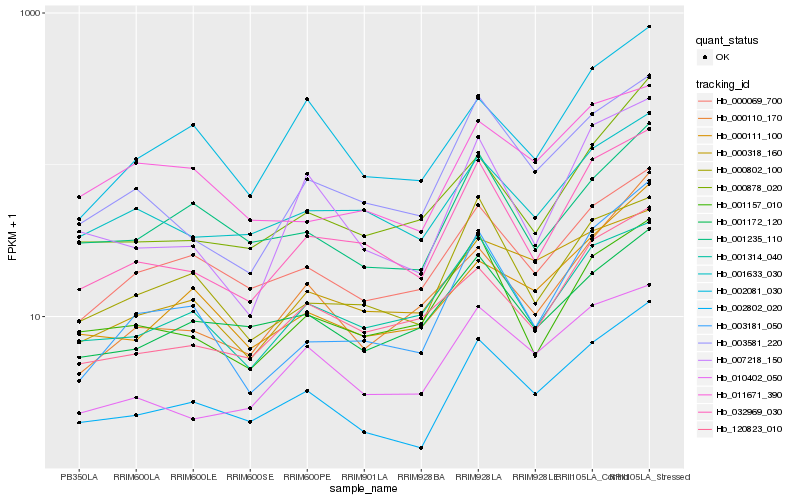
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002802_020 |
0.0 |
- |
- |
PREDICTED: putative auxin efflux carrier component 5 [Jatropha curcas] |
| 2 |
Hb_032969_030 |
0.1173459617 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 3 |
Hb_000069_700 |
0.1232347932 |
- |
- |
hypothetical protein B456_005G003200 [Gossypium raimondii] |
| 4 |
Hb_007218_150 |
0.1238171244 |
- |
- |
RecName: Full=Profilin-3; AltName: Full=Pollen allergen Hev b 8.0201; AltName: Allergen=Hev b 8.0201 [Hevea brasiliensis] |
| 5 |
Hb_000111_100 |
0.1248456465 |
- |
- |
PREDICTED: NADPH:quinone oxidoreductase-like [Camelina sativa] |
| 6 |
Hb_001235_110 |
0.1273972181 |
- |
- |
PREDICTED: vesicle-associated protein 1-2-like [Jatropha curcas] |
| 7 |
Hb_003581_220 |
0.1291916534 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 8 |
Hb_003181_050 |
0.131782934 |
- |
- |
PREDICTED: DNA replication complex GINS protein PSF2 [Jatropha curcas] |
| 9 |
Hb_002081_030 |
0.1378060685 |
- |
- |
unnamed protein product [Coffea canephora] |
| 10 |
Hb_010402_050 |
0.1386571984 |
- |
- |
- |
| 11 |
Hb_001172_120 |
0.1445958445 |
- |
- |
hypothetical protein B456_012G082000 [Gossypium raimondii] |
| 12 |
Hb_000878_020 |
0.144978604 |
- |
- |
PREDICTED: uncharacterized protein LOC105629937 [Jatropha curcas] |
| 13 |
Hb_000802_100 |
0.1457088057 |
- |
- |
PREDICTED: uncharacterized protein LOC105634009 [Jatropha curcas] |
| 14 |
Hb_000110_170 |
0.1464689569 |
- |
- |
PREDICTED: arginine biosynthesis bifunctional protein ArgJ, chloroplastic isoform X1 [Jatropha curcas] |
| 15 |
Hb_120823_010 |
0.1473439623 |
- |
- |
- |
| 16 |
Hb_001633_030 |
0.150327635 |
- |
- |
- |
| 17 |
Hb_001314_040 |
0.1522927352 |
- |
- |
hypothetical protein POPTR_0011s10470g [Populus trichocarpa] |
| 18 |
Hb_011671_390 |
0.1546907696 |
- |
- |
peptide methionine sulfoxide reductase A1-like [Jatropha curcas] |
| 19 |
Hb_001157_010 |
0.1554248234 |
- |
- |
deoxyhypusine synthase, putative [Ricinus communis] |
| 20 |
Hb_000318_160 |
0.1594924786 |
- |
- |
PREDICTED: uncharacterized protein LOC105634239 [Jatropha curcas] |