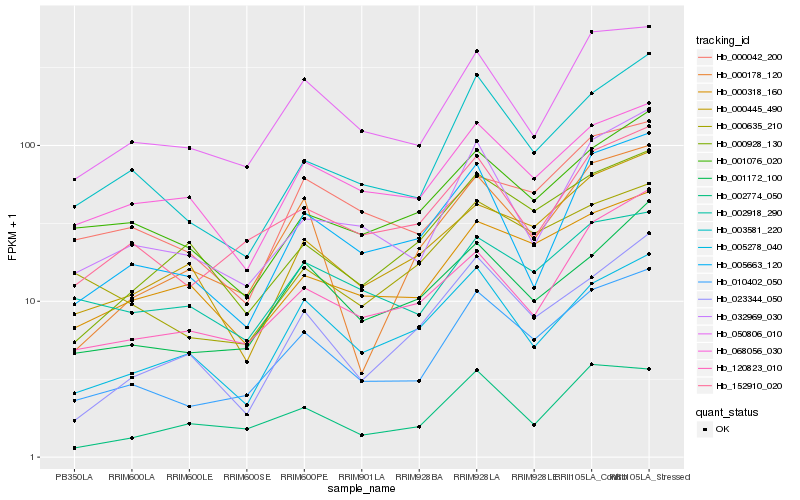
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010402_050 |
0.0 |
- |
- |
- |
| 2 |
Hb_003581_220 |
0.0932056473 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 3 |
Hb_050806_010 |
0.0981487891 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2-17 kDa [Jatropha curcas] |
| 4 |
Hb_152910_020 |
0.1002572676 |
- |
- |
PREDICTED: lactoylglutathione lyase-like [Jatropha curcas] |
| 5 |
Hb_001172_100 |
0.1072832236 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex assembly factor 3-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_032969_030 |
0.1126337733 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 7 |
Hb_000318_160 |
0.1190907239 |
- |
- |
PREDICTED: uncharacterized protein LOC105634239 [Jatropha curcas] |
| 8 |
Hb_001076_020 |
0.1220634865 |
- |
- |
PREDICTED: cytochrome b-c1 complex subunit 8 [Eucalyptus grandis] |
| 9 |
Hb_120823_010 |
0.1222254176 |
- |
- |
- |
| 10 |
Hb_000635_210 |
0.12238846 |
- |
- |
unknown [Populus trichocarpa] |
| 11 |
Hb_068056_030 |
0.1235133008 |
- |
- |
ATP synthase epsilon chain, mitochondrial, putative [Ricinus communis] |
| 12 |
Hb_000445_490 |
0.1241761748 |
- |
- |
- |
| 13 |
Hb_005278_040 |
0.1250957016 |
- |
- |
PREDICTED: sorcin-like isoform X1 [Populus euphratica] |
| 14 |
Hb_023344_050 |
0.1252543738 |
- |
- |
PREDICTED: uncharacterized protein LOC105644474 [Jatropha curcas] |
| 15 |
Hb_002774_050 |
0.1268870586 |
- |
- |
PREDICTED: uncharacterized protein LOC105630601 [Jatropha curcas] |
| 16 |
Hb_005663_120 |
0.1271563435 |
- |
- |
PREDICTED: uncharacterized protein LOC105635265 [Jatropha curcas] |
| 17 |
Hb_000928_130 |
0.1291609704 |
- |
- |
PREDICTED: protein BOLA4, chloroplastic/mitochondrial [Jatropha curcas] |
| 18 |
Hb_000042_200 |
0.1299665084 |
- |
- |
PREDICTED: uncharacterized protein LOC100252136 isoform X1 [Vitis vinifera] |
| 19 |
Hb_002918_290 |
0.1304121405 |
- |
- |
PREDICTED: uncharacterized protein LOC105649583 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000178_120 |
0.130444027 |
- |
- |
PREDICTED: histone H2B [Jatropha curcas] |