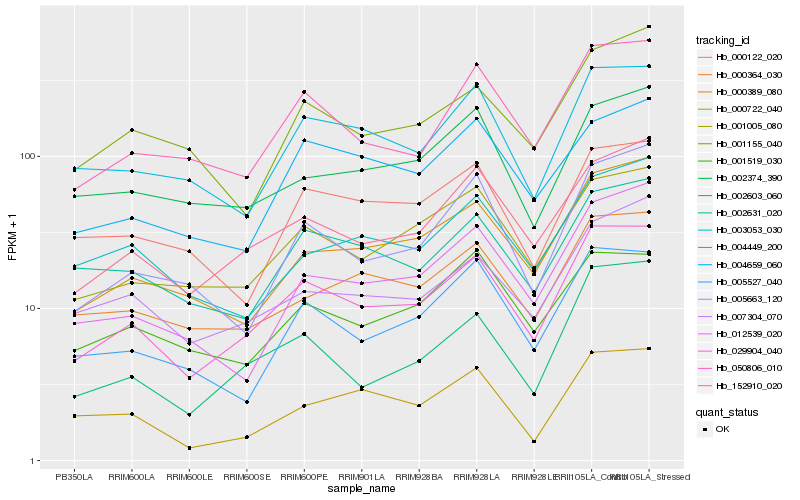
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029904_040 |
0.0 |
- |
- |
putative aspartate aminotransferase protein, partial [Elaeis guineensis] |
| 2 |
Hb_152910_020 |
0.0794400149 |
- |
- |
PREDICTED: lactoylglutathione lyase-like [Jatropha curcas] |
| 3 |
Hb_050806_010 |
0.0887872321 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2-17 kDa [Jatropha curcas] |
| 4 |
Hb_001005_080 |
0.0889614234 |
- |
- |
PREDICTED: cytochrome b-c1 complex subunit Rieske-4, mitochondrial-like [Jatropha curcas] |
| 5 |
Hb_000389_080 |
0.0960100595 |
- |
- |
PREDICTED: uncharacterized protein At4g26485-like [Jatropha curcas] |
| 6 |
Hb_004449_200 |
0.0981210923 |
- |
- |
PREDICTED: nascent polypeptide-associated complex subunit alpha-like protein 1 [Populus euphratica] |
| 7 |
Hb_000364_030 |
0.0996264275 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 8 |
Hb_005527_040 |
0.1000228969 |
- |
- |
PREDICTED: methionine adenosyltransferase 2 subunit beta [Jatropha curcas] |
| 9 |
Hb_002603_060 |
0.1027359261 |
- |
- |
PREDICTED: protein EARLY FLOWERING 4 [Jatropha curcas] |
| 10 |
Hb_002631_020 |
0.1057504201 |
- |
- |
PREDICTED: protein FAM32A-like [Jatropha curcas] |
| 11 |
Hb_007304_070 |
0.1061971765 |
- |
- |
hypoxanthine-guanine phosphoribosyltransferase, putative [Ricinus communis] |
| 12 |
Hb_005663_120 |
0.1101504471 |
- |
- |
PREDICTED: uncharacterized protein LOC105635265 [Jatropha curcas] |
| 13 |
Hb_004659_060 |
0.1121040056 |
- |
- |
RAS-related GTP-binding family protein [Populus trichocarpa] |
| 14 |
Hb_012539_020 |
0.112530237 |
- |
- |
protein binding/ubiquitin-protein ligase 4 [Hevea brasiliensis] |
| 15 |
Hb_001519_030 |
0.1128114059 |
- |
- |
PREDICTED: protein trichome birefringence-like 12 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000122_020 |
0.1148594379 |
- |
- |
PREDICTED: alpha-soluble NSF attachment protein 2 [Jatropha curcas] |
| 17 |
Hb_002374_390 |
0.1149709088 |
- |
- |
vacuolar ATP synthase proteolipid subunit 1, 2, 3, putative [Ricinus communis] |
| 18 |
Hb_000722_040 |
0.1158178858 |
- |
- |
- |
| 19 |
Hb_003053_030 |
0.117929119 |
- |
- |
PREDICTED: transport and Golgi organization 2 homolog [Jatropha curcas] |
| 20 |
Hb_001155_040 |
0.1180012059 |
- |
- |
PREDICTED: cytochrome b-c1 complex subunit 8 [Eucalyptus grandis] |