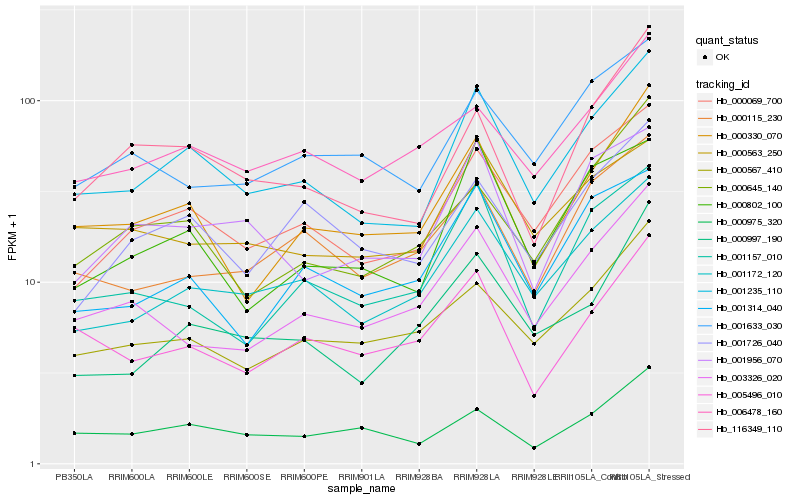
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001235_110 |
0.0 |
- |
- |
PREDICTED: vesicle-associated protein 1-2-like [Jatropha curcas] |
| 2 |
Hb_000069_700 |
0.0821488602 |
- |
- |
hypothetical protein B456_005G003200 [Gossypium raimondii] |
| 3 |
Hb_001172_120 |
0.0875826836 |
- |
- |
hypothetical protein B456_012G082000 [Gossypium raimondii] |
| 4 |
Hb_000330_070 |
0.1005147638 |
- |
- |
PREDICTED: proteasome subunit alpha type-1-B-like [Gossypium raimondii] |
| 5 |
Hb_000802_100 |
0.1094112173 |
- |
- |
PREDICTED: uncharacterized protein LOC105634009 [Jatropha curcas] |
| 6 |
Hb_116349_110 |
0.1144011307 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 25 [Jatropha curcas] |
| 7 |
Hb_000975_320 |
0.1152968749 |
- |
- |
PREDICTED: uncharacterized protein LOC105628218 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000567_410 |
0.1175570697 |
- |
- |
hypothetical protein JCGZ_12784 [Jatropha curcas] |
| 9 |
Hb_006478_160 |
0.1179575473 |
- |
- |
20S proteasome beta subunit D1 [Hevea brasiliensis] |
| 10 |
Hb_001956_070 |
0.1180800991 |
- |
- |
PREDICTED: telomere repeat-binding protein 2 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000563_250 |
0.1202000642 |
- |
- |
PREDICTED: uncharacterized protein LOC105643472 [Jatropha curcas] |
| 12 |
Hb_001314_040 |
0.1216519465 |
- |
- |
hypothetical protein POPTR_0011s10470g [Populus trichocarpa] |
| 13 |
Hb_001726_040 |
0.1216657554 |
- |
- |
hypothetical protein CICLE_v10029230mg [Citrus clementina] |
| 14 |
Hb_001157_010 |
0.1220067557 |
- |
- |
deoxyhypusine synthase, putative [Ricinus communis] |
| 15 |
Hb_005496_010 |
0.1221900973 |
- |
- |
PREDICTED: PXMP2/4 family protein 4 [Jatropha curcas] |
| 16 |
Hb_000115_230 |
0.1222527462 |
- |
- |
PREDICTED: cysteine--tRNA ligase, cytoplasmic-like [Jatropha curcas] |
| 17 |
Hb_000997_190 |
0.1234457055 |
- |
- |
PREDICTED: magnesium transporter MRS2-I-like [Jatropha curcas] |
| 18 |
Hb_003326_020 |
0.1250890974 |
- |
- |
PREDICTED: replication protein A 32 kDa subunit B [Jatropha curcas] |
| 19 |
Hb_000645_140 |
0.1257871795 |
- |
- |
PREDICTED: DNA-directed RNA polymerases II, IV and V subunit 9A [Nelumbo nucifera] |
| 20 |
Hb_001633_030 |
0.1258385437 |
- |
- |
- |