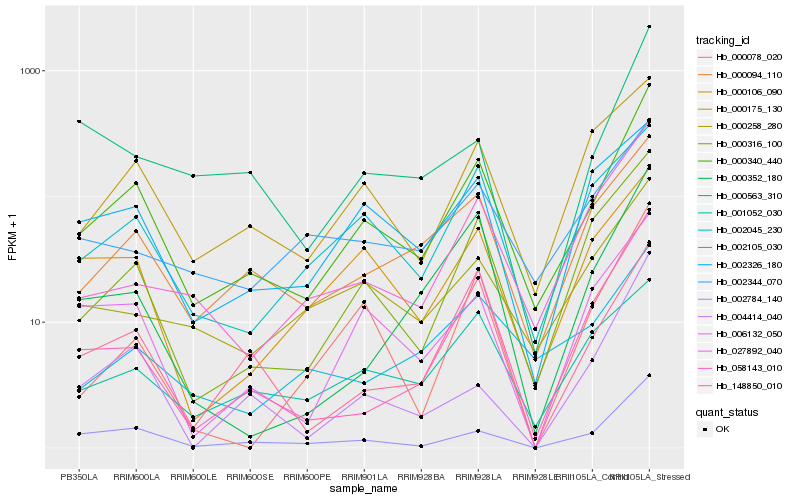
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000340_440 |
0.0 |
- |
- |
PREDICTED: gibberellin receptor GID1B [Jatropha curcas] |
| 2 |
Hb_002784_140 |
0.1385918531 |
- |
- |
glutamate receptor 2 plant, putative [Ricinus communis] |
| 3 |
Hb_000316_100 |
0.1453595043 |
- |
- |
protein with unknown function [Ricinus communis] |
| 4 |
Hb_000094_110 |
0.1554104263 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000175_130 |
0.1645264923 |
- |
- |
PREDICTED: PTI1-like tyrosine-protein kinase At3g15890 [Jatropha curcas] |
| 6 |
Hb_006132_050 |
0.1680125674 |
- |
- |
hypothetical protein EUTSA_v10028860mg [Eutrema salsugineum] |
| 7 |
Hb_148850_010 |
0.1717613952 |
- |
- |
PREDICTED: potassium transporter 5-like [Jatropha curcas] |
| 8 |
Hb_058143_010 |
0.1732224011 |
- |
- |
hypothetical protein JCGZ_01992 [Jatropha curcas] |
| 9 |
Hb_002045_230 |
0.1736452423 |
- |
- |
Ammonium transporter 2 [Theobroma cacao] |
| 10 |
Hb_027892_040 |
0.1763269299 |
- |
- |
PREDICTED: proteasome subunit alpha type-6 [Jatropha curcas] |
| 11 |
Hb_000258_280 |
0.1791775618 |
- |
- |
PREDICTED: probable ran guanine nucleotide release factor [Jatropha curcas] |
| 12 |
Hb_000106_090 |
0.1801341189 |
- |
- |
PREDICTED: uncharacterized protein LOC105634795 [Jatropha curcas] |
| 13 |
Hb_001052_030 |
0.1889771156 |
- |
- |
PREDICTED: protein TIC 20-IV, chloroplastic [Jatropha curcas] |
| 14 |
Hb_002326_180 |
0.1890065083 |
- |
- |
Protein Z, putative [Ricinus communis] |
| 15 |
Hb_002105_030 |
0.1908031401 |
- |
- |
- |
| 16 |
Hb_002344_070 |
0.1915383402 |
- |
- |
PREDICTED: proteasome subunit alpha type-6 [Jatropha curcas] |
| 17 |
Hb_000563_310 |
0.1935069086 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 3 [Hevea brasiliensis] |
| 18 |
Hb_000352_180 |
0.1937139319 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000078_020 |
0.1949653849 |
- |
- |
- |
| 20 |
Hb_004414_040 |
0.1958400853 |
transcription factor |
TF Family: ERF |
AP2 domain-containing transcription factor family protein [Populus trichocarpa] |