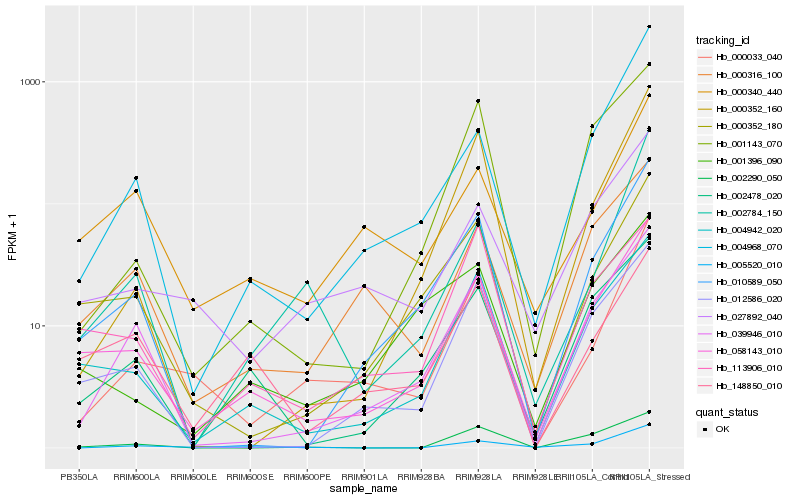
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_058143_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_01992 [Jatropha curcas] |
| 2 |
Hb_004942_020 |
0.0870516755 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 3 |
Hb_000352_180 |
0.1248717957 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_010589_050 |
0.1305827095 |
- |
- |
- |
| 5 |
Hb_012586_020 |
0.13529799 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002478_020 |
0.1374462701 |
- |
- |
hypothetical protein POPTR_0010s19120g [Populus trichocarpa] |
| 7 |
Hb_000316_100 |
0.1557463181 |
- |
- |
protein with unknown function [Ricinus communis] |
| 8 |
Hb_039946_010 |
0.1564342085 |
- |
- |
hypothetical protein POPTR_0018s09200g [Populus trichocarpa] |
| 9 |
Hb_027892_040 |
0.1640717015 |
- |
- |
PREDICTED: proteasome subunit alpha type-6 [Jatropha curcas] |
| 10 |
Hb_148850_010 |
0.1643533136 |
- |
- |
PREDICTED: potassium transporter 5-like [Jatropha curcas] |
| 11 |
Hb_004968_070 |
0.1667163987 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000340_440 |
0.1732224011 |
- |
- |
PREDICTED: gibberellin receptor GID1B [Jatropha curcas] |
| 13 |
Hb_113906_010 |
0.1761133363 |
- |
- |
hypothetical protein JCGZ_18857 [Jatropha curcas] |
| 14 |
Hb_001396_090 |
0.1821648802 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 75 [Jatropha curcas] |
| 15 |
Hb_001143_070 |
0.1842551353 |
- |
- |
Acyl-CoA N-acyltransferases (NAT) superfamily protein isoform 1 [Theobroma cacao] |
| 16 |
Hb_000033_040 |
0.1878406483 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000352_160 |
0.1955194889 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002290_050 |
0.1956409805 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002784_150 |
0.1968466867 |
- |
- |
hypothetical protein POPTR_0001s06620g [Populus trichocarpa] |
| 20 |
Hb_005520_010 |
0.1972030666 |
- |
- |
hypothetical protein JCGZ_09913 [Jatropha curcas] |