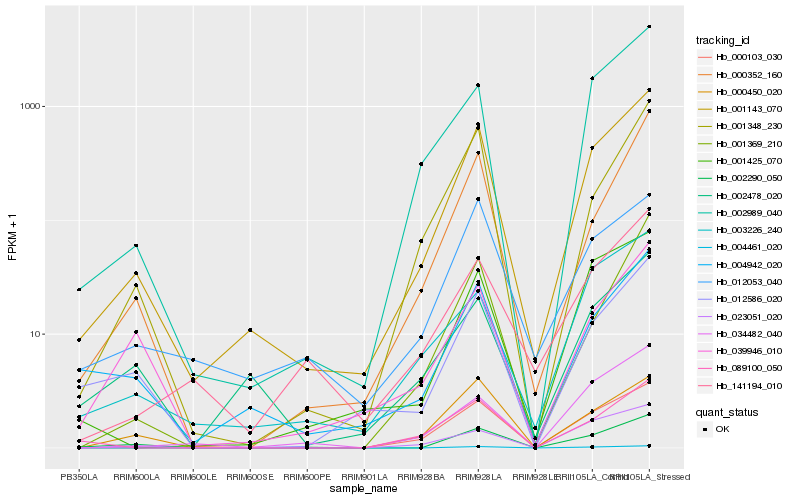
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001143_070 |
0.0 |
- |
- |
Acyl-CoA N-acyltransferases (NAT) superfamily protein isoform 1 [Theobroma cacao] |
| 2 |
Hb_002989_040 |
0.123710127 |
- |
- |
hypothetical protein 1 [Hevea brasiliensis] |
| 3 |
Hb_000103_030 |
0.130175638 |
- |
- |
hypothetical protein VITISV_021700 [Vitis vinifera] |
| 4 |
Hb_001348_230 |
0.1399552663 |
transcription factor |
TF Family: TRAF |
protein with unknown function [Ricinus communis] |
| 5 |
Hb_002290_050 |
0.1404987917 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_004942_020 |
0.1405291483 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 7 |
Hb_000352_160 |
0.1445255586 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001425_070 |
0.1465315609 |
- |
- |
- |
| 9 |
Hb_012586_020 |
0.1491681038 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 isoform X1 [Jatropha curcas] |
| 10 |
Hb_003226_240 |
0.1496923468 |
- |
- |
PREDICTED: caffeoylshikimate esterase [Jatropha curcas] |
| 11 |
Hb_004461_020 |
0.1531337875 |
- |
- |
PREDICTED: uncharacterized protein LOC104105697 [Nicotiana tomentosiformis] |
| 12 |
Hb_141194_010 |
0.1559356842 |
- |
- |
PREDICTED: protein ZINC INDUCED FACILITATOR 1-like isoform X2 [Populus euphratica] |
| 13 |
Hb_012053_040 |
0.1585353717 |
- |
- |
PREDICTED: putative phospholipid:diacylglycerol acyltransferase 2 [Jatropha curcas] |
| 14 |
Hb_002478_020 |
0.1603811398 |
- |
- |
hypothetical protein POPTR_0010s19120g [Populus trichocarpa] |
| 15 |
Hb_001369_210 |
0.1638174384 |
transcription factor |
TF Family: C2H2 |
hypothetical protein JCGZ_26269 [Jatropha curcas] |
| 16 |
Hb_023051_020 |
0.1639787032 |
- |
- |
- |
| 17 |
Hb_034482_040 |
0.1644455097 |
- |
- |
aspartate kinase, partial [Francisella tularensis] |
| 18 |
Hb_089100_050 |
0.1646651662 |
- |
- |
- |
| 19 |
Hb_039946_010 |
0.1652546762 |
- |
- |
hypothetical protein POPTR_0018s09200g [Populus trichocarpa] |
| 20 |
Hb_000450_020 |
0.1666183338 |
- |
- |
conserved hypothetical protein [Ricinus communis] |