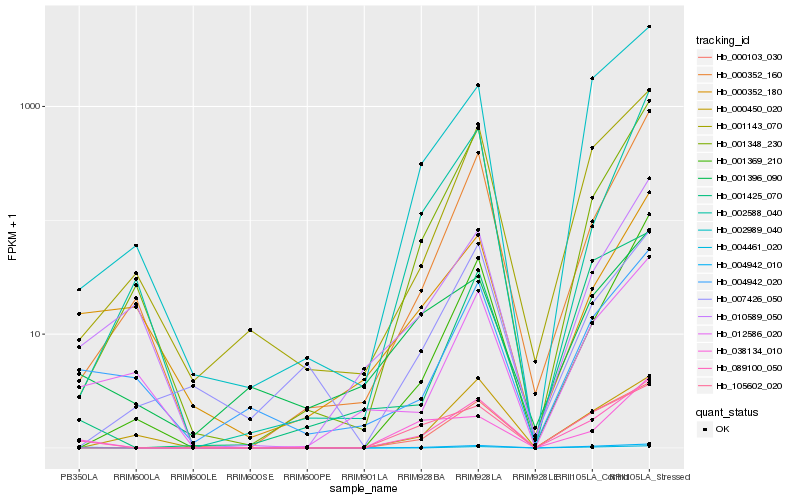
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_089100_050 |
0.0 |
- |
- |
- |
| 2 |
Hb_004942_010 |
0.1287349879 |
- |
- |
hypothetical protein AMTR_s00057p00201180 [Amborella trichopoda] |
| 3 |
Hb_000103_030 |
0.1353917787 |
- |
- |
hypothetical protein VITISV_021700 [Vitis vinifera] |
| 4 |
Hb_001348_230 |
0.1565862445 |
transcription factor |
TF Family: TRAF |
protein with unknown function [Ricinus communis] |
| 5 |
Hb_105602_020 |
0.1642164472 |
- |
- |
PREDICTED: 60S ribosomal protein L19-1-like [Populus euphratica] |
| 6 |
Hb_001143_070 |
0.1646651662 |
- |
- |
Acyl-CoA N-acyltransferases (NAT) superfamily protein isoform 1 [Theobroma cacao] |
| 7 |
Hb_002989_040 |
0.1726680414 |
- |
- |
hypothetical protein 1 [Hevea brasiliensis] |
| 8 |
Hb_004942_020 |
0.1805996184 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 9 |
Hb_001369_210 |
0.1869584351 |
transcription factor |
TF Family: C2H2 |
hypothetical protein JCGZ_26269 [Jatropha curcas] |
| 10 |
Hb_000352_160 |
0.1874909363 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_012586_020 |
0.189596698 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 isoform X1 [Jatropha curcas] |
| 12 |
Hb_038134_010 |
0.1910952338 |
- |
- |
- |
| 13 |
Hb_001396_090 |
0.193484626 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 75 [Jatropha curcas] |
| 14 |
Hb_010589_050 |
0.1975931003 |
- |
- |
- |
| 15 |
Hb_002588_040 |
0.1996687011 |
- |
- |
Pathogenesis-related protein 1 [Theobroma cacao] |
| 16 |
Hb_000450_020 |
0.204058051 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_004461_020 |
0.2083719588 |
- |
- |
PREDICTED: uncharacterized protein LOC104105697 [Nicotiana tomentosiformis] |
| 18 |
Hb_001425_070 |
0.2095360632 |
- |
- |
- |
| 19 |
Hb_000352_180 |
0.2099491474 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_007426_050 |
0.2108799662 |
transcription factor |
TF Family: Tify |
JAZ8 [Hevea brasiliensis] |