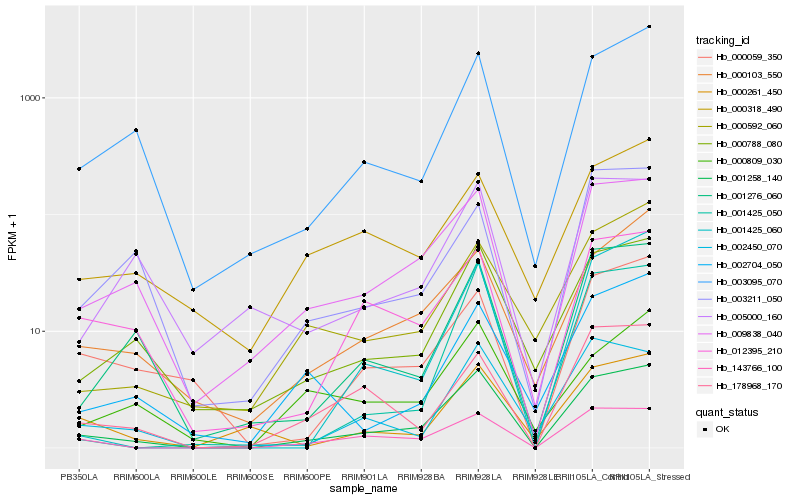
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001258_140 |
0.0 |
- |
- |
- |
| 2 |
Hb_009838_040 |
0.1287178194 |
- |
- |
hypothetical protein JCGZ_12866 [Jatropha curcas] |
| 3 |
Hb_143766_100 |
0.1409259079 |
- |
- |
PREDICTED: uncharacterized protein LOC105637800 [Jatropha curcas] |
| 4 |
Hb_000788_080 |
0.1530219234 |
- |
- |
PREDICTED: uncharacterized protein LOC105629703 isoform X1 [Jatropha curcas] |
| 5 |
Hb_003095_070 |
0.1553059667 |
- |
- |
hypothetical protein POPTR_0009s11230g [Populus trichocarpa] |
| 6 |
Hb_012395_210 |
0.1629355395 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001276_060 |
0.1631947437 |
- |
- |
hypothetical protein POPTR_0015s00420g [Populus trichocarpa] |
| 8 |
Hb_002450_070 |
0.1687317445 |
- |
- |
(S)-N-methylcoclaurine 3'-hydroxylase isozyme, putative [Ricinus communis] |
| 9 |
Hb_000103_550 |
0.1691335929 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit gamma 2 isoform X1 [Jatropha curcas] |
| 10 |
Hb_178968_170 |
0.1731215105 |
- |
- |
PREDICTED: protein SUPPRESSOR OF GENE SILENCING 3 homolog [Jatropha curcas] |
| 11 |
Hb_001425_060 |
0.1779723492 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000261_450 |
0.1817430896 |
- |
- |
Pectinesterase U1 precursor, putative [Ricinus communis] |
| 13 |
Hb_001425_050 |
0.1817869243 |
- |
- |
- |
| 14 |
Hb_000059_350 |
0.1824246942 |
- |
- |
glutamate dehydrogenase, putative [Ricinus communis] |
| 15 |
Hb_002704_050 |
0.1887317298 |
- |
- |
cysteine-type peptidase, putative [Ricinus communis] |
| 16 |
Hb_000592_060 |
0.1903869607 |
- |
- |
PREDICTED: ras-related protein RABA6a [Populus euphratica] |
| 17 |
Hb_005000_160 |
0.1920813798 |
- |
- |
PREDICTED: uncharacterized protein LOC105637882 [Jatropha curcas] |
| 18 |
Hb_000809_030 |
0.19321892 |
desease resistance |
Gene Name: RPW8 |
PREDICTED: probable disease resistance protein At5g66900 [Prunus mume] |
| 19 |
Hb_003211_050 |
0.1940161338 |
- |
- |
PREDICTED: RNA-binding protein 24-B isoform X1 [Jatropha curcas] |
| 20 |
Hb_000318_490 |
0.1943417023 |
- |
- |
PREDICTED: mitochondrial import receptor subunit TOM6 homolog [Jatropha curcas] |